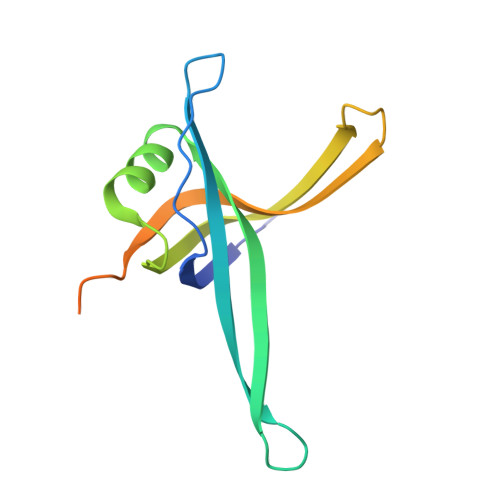
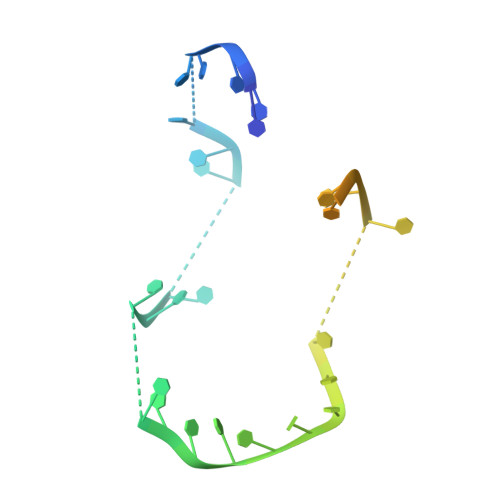
Single-Stranded DNA-Binding Protein Complex from Helicobacter Pylori Suggests an Ssdna-Binding Surface.
Chan, K.-W., Lee, Y.-J., Wang, C.-H., Huang, H., Sun, Y.-J.(2009) J Mol Biology 388: 508
- PubMed: 19285993
- DOI: https://doi.org/10.1016/j.jmb.2009.03.022
- Primary Citation of Related Structures:
2VW9 - PubMed Abstract:
Single-stranded DNA (ssDNA)-binding protein (SSB) plays an important role in DNA replication, recombination, and repair. SSB consists of an N-terminal ssDNA-binding domain with an oligonucleotide/oligosaccharide binding fold and a flexible C-terminal tail involved in protein-protein interactions. SSB from Helicobacter pylori (HpSSB) was isolated, and the ssDNA-binding characteristics of HpSSB were analyzed by fluorescence titration and electrophoretic mobility shift assay. Tryptophan fluorescence quenching was measured as 61%, and the calculated cooperative affinity was 5.4x10(7) M(-1) with an ssDNA-binding length of 25-30 nt. The crystal structure of the C-terminally truncated protein (HpSSBc) in complex with 35-mer ssDNA [HpSSBc-(dT)(35)] was determined at a resolution of 2.3 A. The HpSSBc monomer folds as an oligonucleotide/oligosaccharide binding fold with a Y-shaped conformation. The ssDNA wrapped around the HpSSBc tetramer through a continuous binding path comprising five essential aromatic residues and a positively charged surface formed by numerous basic residues.
- Institute of Bioinformatics and Structural Biology, National Tsing Hua University, Hsinchu 300, Taiwan, ROC.
Organizational Affiliation: