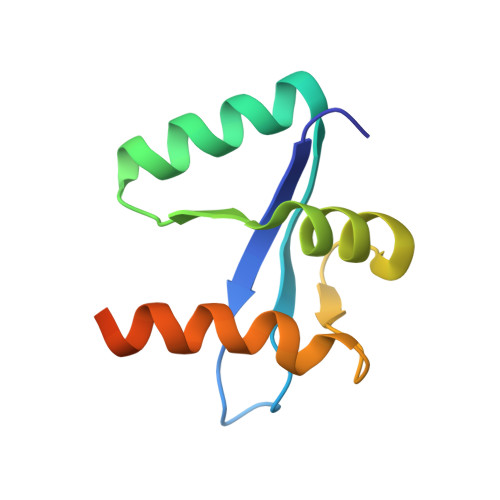
Crystal Structure of Spa40, the Specificity Switch for the Shigella Flexneri Type III Secretion System
Deane, J.E., Graham, S.C., Mitchell, E.P., Flot, D., Johnson, S., Lea, S.M.(2008) Mol Microbiol 69: 267
- PubMed: 18485071
- DOI: https://doi.org/10.1111/j.1365-2958.2008.06293.x
- Primary Citation of Related Structures:
2VT1 - PubMed Abstract:
The pathogenic bacterium Shigella flexneri uses a type III secretion system to inject virulence factors from the bacterial cytosol directly into host cells. The machinery that identifies secretion substrates and controls the export of extracellular components and effector proteins consists of several inner-membrane and cytoplasmic proteins. One of the inner membrane components, Spa40, belongs to a family of proteins proposed to regulate the switching of substrate specificity of the export apparatus. We show that Spa40 is cleaved within the strictly conserved amino acid sequence NPTH and substitution of the proposed autocatalytic residue abolishes cleavage. Here we also report the crystal structure of the cytoplasmic complex Spa40(C) and compare it with the recent structures of the homologues from Escherichia coli and Salmonella typhimurium. These structures reveal the tight association of the cleaved fragments and show that the conserved NPTH sequence lies on a loop which, when cleaved, swings away from the catalytic N257 residue, resulting in different surface features in this region. This structural rearrangement suggests a mechanism by which non-cleaving forms of these proteins interfere with correct substrate switching of the apparatus.
- Sir William Dunn School of Pathology, South Parks Rd, University of Oxford, OX1 3RE, UK.
Organizational Affiliation: