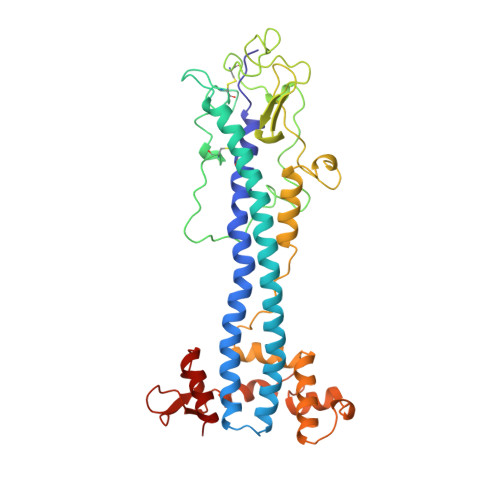
A structural motif in the variant surface glycoproteins of Trypanosoma brucei.
Blum, M.L., Down, J.A., Gurnett, A.M., Carrington, M., Turner, M.J., Wiley, D.C.(1993) Nature 362: 603-609
- PubMed: 8464512
- DOI: https://doi.org/10.1038/362603a0
- Primary Citation of Related Structures:
2VSG - PubMed Abstract:
The variable domain of the trypanosome variant surface glycoprotein (VSG) ILTat 1.24 has been shown by X-ray crystallography to resemble closely the structures of VSG MITat 1.2, despite their low sequence similarity. Specific structural features of these VSGs, including substitution of carbohydrate for an alpha-helix, can be found in other VSG sequences. Thus antigenic variation in trypanosomes is accomplished by sequence variation, not gross structural alteration; the extensive sequence differences among VSGs may be required for another reason, such as the avoidance of recognition by helper T cells. Additionally, VSG sequences are found to define families, within a VSG superfamily, which have evolved in the trypanosome genome.
- Department of Biochemistry and Molecular Biology, Harvard University, Cambridge, Massachusetts.
Organizational Affiliation: