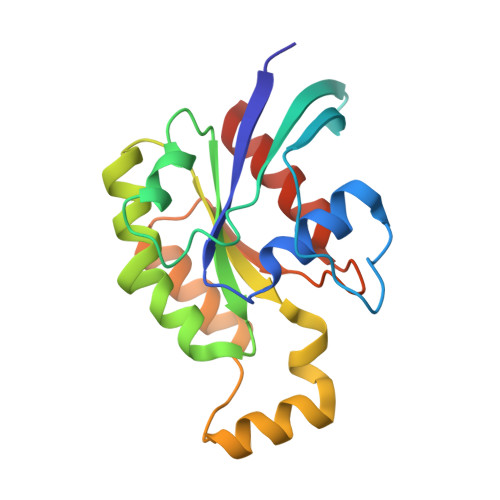
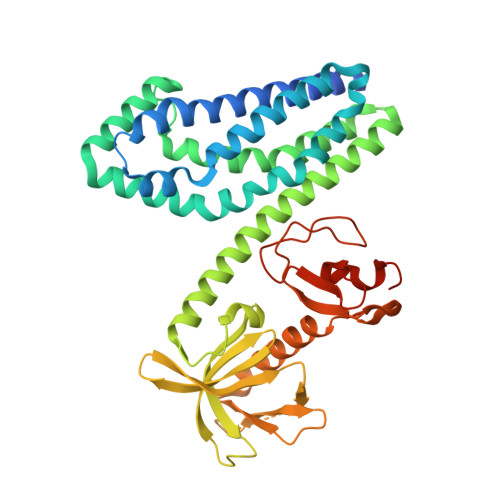
Crucial Structural Role for the Ph and C1 Domains of the Vav1 Exchange Factor.
Rapley, J., Tybulewicz, V., Rittinger, K.(2008) EMBO Rep 9: 655
- PubMed: 18511940
- DOI: https://doi.org/10.1038/embor.2008.80
- Primary Citation of Related Structures:
2VRW - PubMed Abstract:
The Vav family of proteins are guanine nucleotide exchange factors (GEFs) for the Rho family of GTPases, which regulate various cellular functions, including T-cell activation. They contain a catalytic Dbl homology (DH) domain that is invariably followed by a pleckstrin homology (PH) domain, which is often required for catalytic activity. Vav proteins are the first GEFs for which an additional C1 domain is required for full biological activity. Here, we present the structure of a Vav1 fragment comprising the DH-PH-C1 domains bound to Rac1. This structure shows that the PH and C1 domains form a single structural unit that packs against the carboxy-terminal helix of the DH domain to stabilize its conformation and to promote nucleotide exchange. In contrast to previous reports, this structure shows that there are no direct contacts between the GTPase and C1 domain but instead suggests new mechanisms for the regulation of Vav1 activity.
- Division of Molecular Structure, and National Institute for Medical Research, The Ridgeway, London NW7 1AA, UK.
Organizational Affiliation: