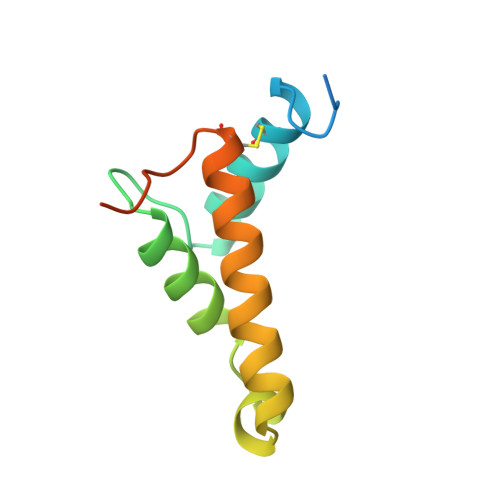
A Putative Alpha-Helical Porin from Corynebacterium Glutamicum.
Ziegler, K., Benz, R., Schulz, G.E.(2008) J Mol Biology 379: 482
- PubMed: 18462756
- DOI: https://doi.org/10.1016/j.jmb.2008.04.017
- Primary Citation of Related Structures:
2VQG, 2VQH, 2VQK, 2VQL - PubMed Abstract:
The cell wall of Corynebacterium glutamicum contains a mycolic acid layer, which is a protective nonpolar barrier similar to the outer membrane of Gram-negative bacteria. The exchange of material across this barrier requires porins. Porin B (PorB) is one of them. Recombinant PorB has been produced in Escherichia coli, purified, crystallized and analyzed by X-ray diffraction, yielding 16 independent molecular structures in four different crystal forms at resolutions up to 1.8 A. All 16 molecules have the same globular core, which consists of 70 residues forming four alpha-helices tied together by a disulfide bridge. The 16 structures vary greatly with respect to the 29 residues in the N- and C-terminal extensions. Since corynebacteria belong to the group of mycolata that includes some prominent human pathogens, the observed structure may be of medical relevance. Due to the clearly established solid structure of the core, the native porin has to be oligomeric, and the reported structure is one of the subunits. An alpha-helical porin in a bacterial outer envelope is surprising because all presently known structures of such porins consist of beta-barrels. Since none of the four crystal packing arrangements was compatible with an oligomeric membrane channel, we constructed a model of such an oligomer that was consistent with all available data of native PorB. The proposed model is based on the required polar interior and nonpolar exterior of the porin, on a recurring crystal packing contact around a 2-fold axis, on the assumption of a simple C(n) symmetry (a symmetric arrangement around an n-fold axis), on the experimentally established electric conductivity and anion selectivity and on the generally observed shape of porin channels.
- Institut für Organische Chemie und Biochemie, Albert-Ludwigs-Universität, Albertstr. 21, 79104 Freiburg im Breisgau, Germany.
Organizational Affiliation: