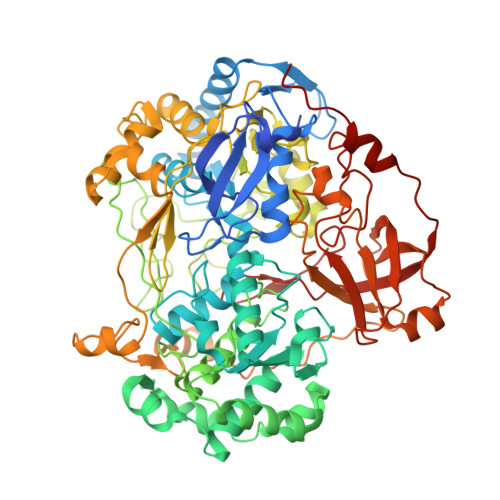
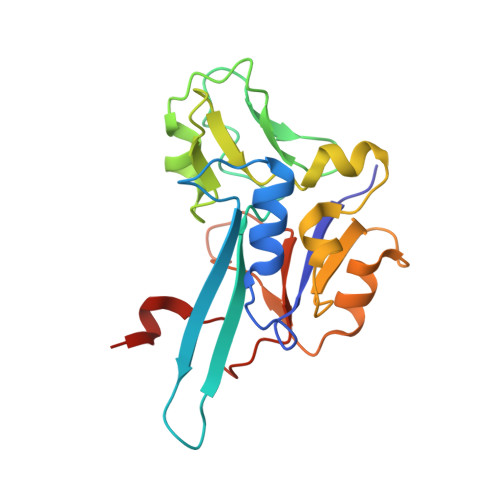
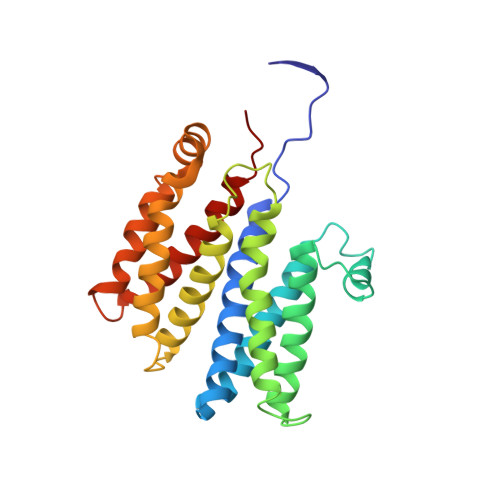
Molecular Mechanism of Energy Conservation in Polysulfide Respiration.
Jormakka, M., Yokoyama, K., Yano, T., Tamakoshi, M., Akimoto, S., Shimamura, T., Curmi, P., Iwata, S.(2008) Nat Struct Mol Biol 15: 730
- PubMed: 18536726
- DOI: https://doi.org/10.1038/nsmb.1434
- Primary Citation of Related Structures:
2VPW, 2VPX, 2VPY, 2VPZ - PubMed Abstract:
Bacterial polysulfide reductase (PsrABC) is an integral membrane protein complex responsible for quinone-coupled reduction of polysulfide, a process important in extreme environments such as deep-sea vents and hot springs. We determined the structure of polysulfide reductase from Thermus thermophilus at 2.4-A resolution, revealing how the PsrA subunit recognizes and reduces its unique polyanionic substrate. The integral membrane subunit PsrC was characterized using the natural substrate menaquinone-7 and inhibitors, providing a comprehensive representation of a quinone binding site and revealing the presence of a water-filled cavity connecting the quinone binding site on the periplasmic side to the cytoplasm. These results suggest that polysulfide reductase could be a key energy-conserving enzyme of the T. thermophilus respiratory chain, using polysulfide as the terminal electron acceptor and pumping protons across the membrane via a previously unknown mechanism.
- Department of Biophysics, University of New South Wales, Barker Street, Sydney, New South Wales 2052, Australia. m.jormakka@centenary.org.au
Organizational Affiliation: