Crystal Structure of a Cellulosomal Family 3 Carbohydrate Esterase from Clostridium Thermocellum Provides Insights Into the Mechanism of Substrate Recognition
Correia, M.A.S., Prates, J.A.M., Bras, J., Fontes, C.M.G.A., Newman, J.A., Lewis, R.J., Gilbert, H.J., Flint, J.E.(2008) J Mol Biology 379: 64
- PubMed: 18436237
- DOI: https://doi.org/10.1016/j.jmb.2008.03.037
- Primary Citation of Related Structures:
2VPT - PubMed Abstract:
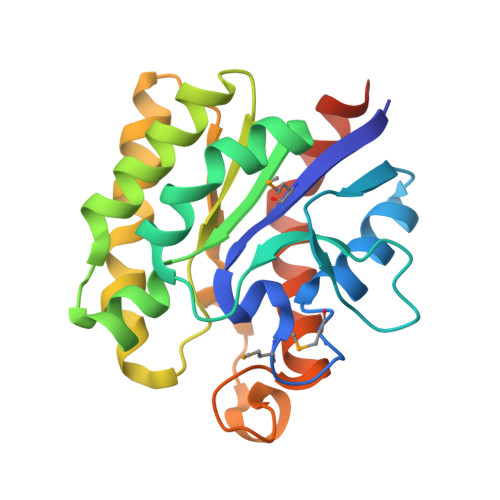
The microbial degradation of the plant cell wall is of increasing industrial significance, exemplified by the interest in generating biofuels from plant cell walls. The majority of plant cell-wall polysaccharides are acetylated, and removal of the acetyl groups through the action of carbohydrate esterases greatly increases the efficiency of polysaccharide saccharification. Enzymes in carbohydrate esterase family 3 (CE3) are common in plant cell wall-degrading microorganisms but there is a paucity of structural and biochemical information on these biocatalysts. Clostridium thermocellum contains a single CE3 enzyme, CtCes3, which comprises two highly homologous (97% sequence identity) catalytic modules appended to a C-terminal type I dockerin that targets the esterase into the cellulosome, a large protein complex that catalyses plant cell wall degradation. Here, we report the crystal structure and biochemical properties of the N-terminal catalytic module (CtCes3-1) of CtCes3. The enzyme is a thermostable acetyl-specific esterase that exhibits a strong preference for acetylated xylan. CtCes3-1 displays an alpha/beta hydrolase fold that contains a central five-stranded parallel twisted beta-sheet flanked by six alpha-helices. In addition, the enzyme contains a canonical catalytic triad in which Ser44 is the nucleophile, His208 is the acid-base and Asp205 modulates the basic nature of the histidine. The acetate moiety is accommodated in a hydrophobic pocket and the negative charge of the tetrahedral transition state is stabilized through hydrogen bonds with the backbone N of Ser44 and Gly95 and the side-chain amide of Asn124.
- CIISA - Faculdade de Medicina Veterinária, Universidade Técnica de Lisboa, Avenida da Universidade Técnica, 1300-477 Lisboa, Portugal.
Organizational Affiliation: