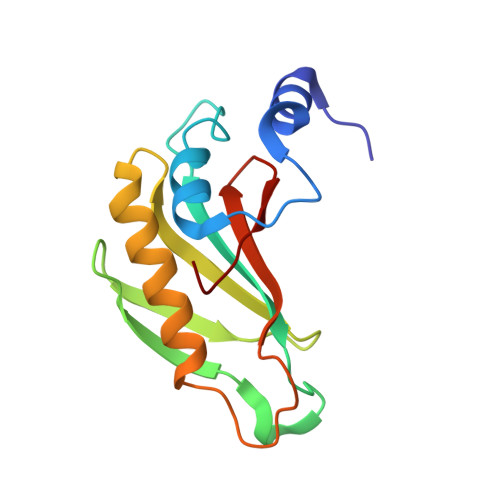
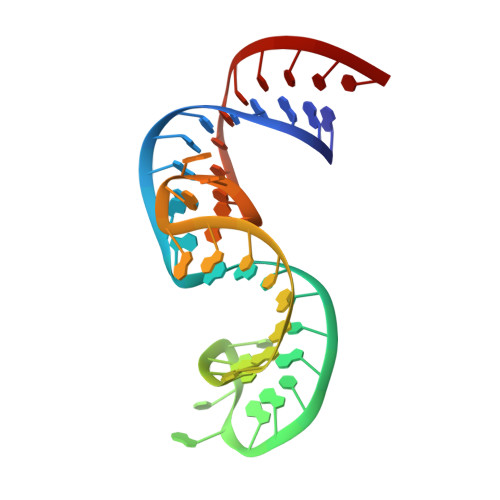
Domain II of Thermus thermophilus ribosomal protein L1 hinders recognition of its mRNA.
Tishchenko, S., Kljashtorny, V., Kostareva, O., Nevskaya, N., Nikulin, A., Gulak, P., Piendl, W., Garber, M., Nikonov, S.(2008) J Mol Biology 383: 301-305
- PubMed: 18778715
- DOI: https://doi.org/10.1016/j.jmb.2008.08.058
- Primary Citation of Related Structures:
2VPL - PubMed Abstract:
The two-domain ribosomal protein L1 has a dual function as a primary rRNA-binding ribosomal protein and as a translational repressor that binds its own mRNA. Here, we report the crystal structure of a complex between the isolated domain I of L1 from the bacterium Thermus thermophilus and a specific mRNA fragment from Methanoccocus vannielii. In parallel, we report kinetic characteristics measured for complexes formed by intact TthL1 and its domain I with the specific mRNA fragment. Although, there is a close similarity between the RNA-protein contact regions in both complexes, the association rate constant is higher in the case of the complex formed by the isolated domain I. This finding demonstrates that domain II hinders mRNA recognition by the intact TthL1.
- Institute of Protein Research, Russian Academy of Sciences, 142290 Pushchino, Moscow region, Russia.
Organizational Affiliation: