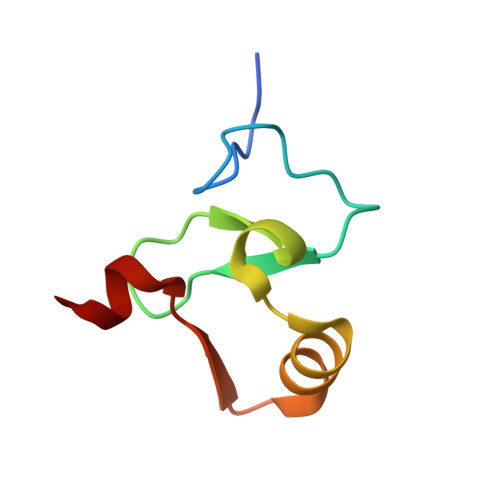
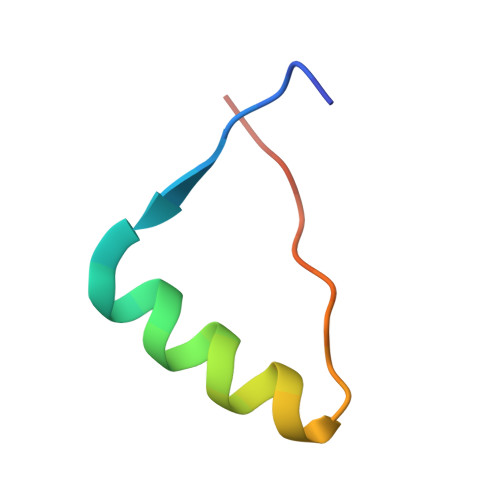
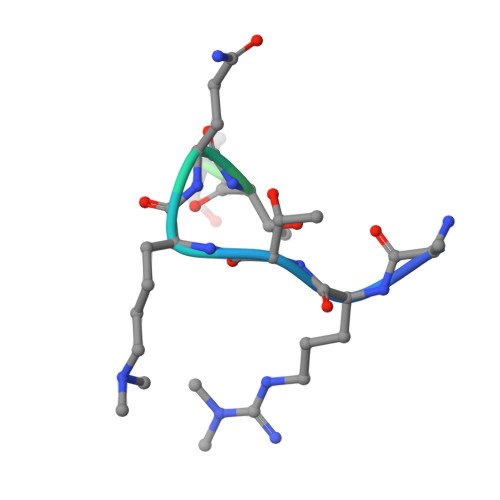
Decoding of Methylated Histone H3 Tail by the Pygo- Bcl9 Wnt Signaling Complex.
Fiedler, M., Sanchez-Barrena, M.J., Nekrasov, M., Mieszczanek, J., Rybin, V., Muller, J., Evans, P., Bienz, M.(2008) Mol Cell 30: 507
- PubMed: 18498752
- DOI: https://doi.org/10.1016/j.molcel.2008.03.011
- Primary Citation of Related Structures:
2VP7, 2VPB, 2VPD, 2VPE, 2VPG - PubMed Abstract:
Pygo and BCL9/Legless transduce the Wnt signal by promoting the transcriptional activity of beta-catenin/Armadillo in normal and malignant cells. We show that human and Drosophila Pygo PHD fingers associate with their cognate HD1 domains from BCL9/Legless to bind specifically to the histone H3 tail methylated at lysine 4 (H3K4me). The crystal structures of ternary complexes between PHD, HD1, and two different H3K4me peptides reveal a unique mode of histone tail recognition: efficient histone binding requires HD1 association, and the PHD-HD1 complex binds preferentially to H3K4me2 while displaying insensitivity to methylation of H3R2. Therefore, this is a prime example of histone tail binding by a PHD finger (of Pygo) being modulated by a cofactor (BCL9/Legless). Rescue experiments in Drosophila indicate that Wnt signaling outputs depend on histone decoding. The specificity of this process provided by the Pygo-BCL9/Legless complex suggests that this complex facilitates an early step in the transition from gene silence to Wnt-induced transcription.
- MRC Laboratory of Molecular Biology, Hills Road, Cambridge CB2 0QH, UK.
Organizational Affiliation: