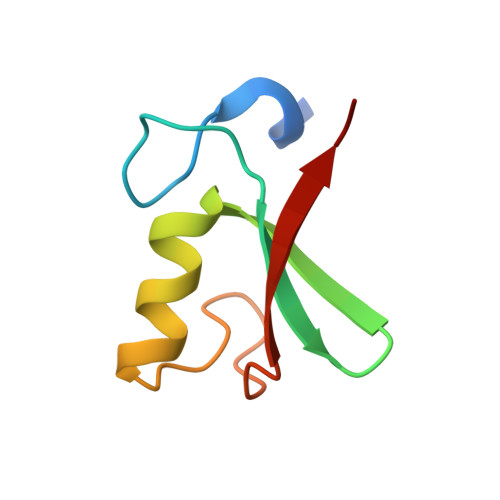
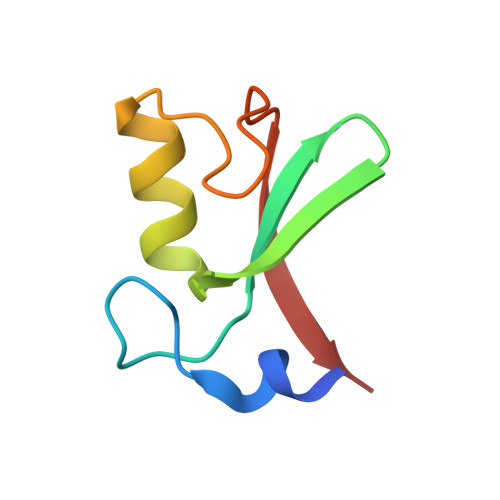
Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans.
Linke, K., Mace, P.D., Smith, C.A., Vaux, D.L., Silke, J., Day, C.L.(2008) Cell Death Differ 15: 841-848
- PubMed: 18219319
- DOI: https://doi.org/10.1038/sj.cdd.4402309
- Primary Citation of Related Structures:
2VJE, 2VJF - PubMed Abstract:
MDM2, a ubiquitin E3-ligase of the RING family, has a key role in regulating p53 abundance. During normal non-stress conditions p53 is targeted for degradation by MDM2. MDM2 can also target itself and MDMX for degradation. MDMX is closely related to MDM2 but the RING domain of MDMX does not possess intrinsic E3-ligase activity. Instead, MDMX regulates p53 abundance by modulating the levels and activity of MDM2. Dimerization, mediated by the conserved C-terminal RING domains of both MDM2 and MDMX, is critical to this activity. Here we report the crystal structure of the MDM2/MDMX RING domain heterodimer and map residues required for functional interaction with the E2 (UbcH5b). In both MDM2 and MDMX residues C-terminal to the RING domain have a key role in dimer formation. In addition we show that these residues are part of an extended surface that is essential for ubiquitylation in trans. This study provides a molecular basis for understanding how heterodimer formation leads to stabilization of MDM2, yet degradation of p53, and suggests novel targets for therapeutic intervention.
- Department of Biochemistry, University of Otago, Dunedin, New Zealand.
Organizational Affiliation: