Solitary and Repetitive Binding Motifs for the Ap2 Complex {Alpha}-Appendage in Amphiphysin and Other Accessory Proteins.
Olesen, L.E., Ford, M.G.J., Schmid, E.M., Vallis, Y., Madan Babu, M., Li, P.H., Mills, I.G., Mcmahon, H.T., Praefcke, G.J.K.(2008) J Biological Chem 283: 5099
- PubMed: 17986441
- DOI: https://doi.org/10.1074/jbc.M708621200
- Primary Citation of Related Structures:
2VJ0 - PubMed Abstract:
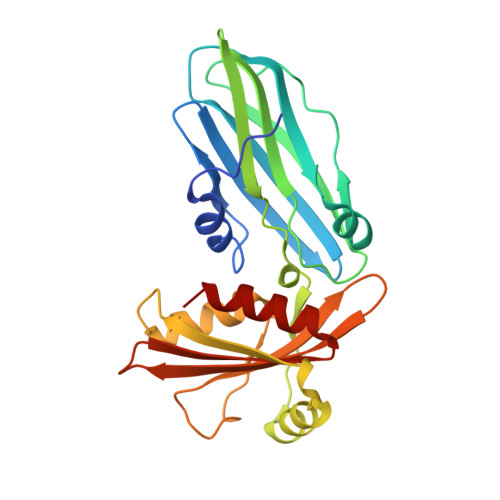
Adaptor protein (AP) complexes bind to transmembrane proteins destined for internalization and to membrane lipids, so linking cargo to the accessory internalization machinery. This machinery interacts with the appendage domains of APs, which have platform and beta-sandwich subdomains, forming the binding surfaces for interacting proteins. Proteins that interact with the subdomains do so via short motifs, usually found in regions of low structural complexity of the interacting proteins. So far, up to four motifs have been identified that bind to and partially compete for at least two sites on each of the appendage domains of the AP2 complex. Motifs in individual accessory proteins, their sequential arrangement into motif domains, and partial competition for binding sites on the appendage domains coordinate the formation of endocytic complexes in a temporal and spatial manner. In this work, we examine the dominant interaction sequence in amphiphysin, a synapse-enriched accessory protein, which generates membrane curvature and recruits the scission protein dynamin to the necks of coated pits, for the platform subdomain of the alpha-appendage. The motif domain of amphiphysin1 contains one copy of each of a DX(F/W) and FXDXF motif. We find that the FXDXF motif is the main determinant for the high affinity interaction with the alpha-adaptin appendage. We describe the optimal sequence of the FXDXF motif using thermodynamic and structural data and show how sequence variation controls the affinities of these motifs for the alpha-appendage.
- Laboratory of Molecular Biology, Medical Research Council, Neurobiology Division, Hills Road, Cambridge CB2 2QH, United Kingdom.
Organizational Affiliation: