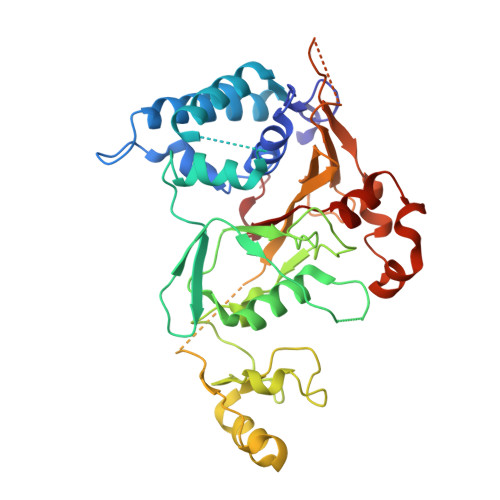
The Structure of the Cyld Usp Domain Explains its Specificity for Lys63-Linked Polyubiquitin and Reveals a B-Box Module
Komander, D., Lord, C.J., Scheel, H., Swift, S., Hofmann, K., Ashworth, A., Barford, D.(2008) Mol Cell Biol 29: 451
- PubMed: 18313383
- DOI: https://doi.org/10.1016/j.molcel.2007.12.018
- Primary Citation of Related Structures:
2VHF - PubMed Abstract:
The tumor suppressor CYLD antagonizes NF-kappaB and JNK signaling by disassembly of Lys63-linked ubiquitin chains synthesized in response to cytokine stimulation. Here we describe the crystal structure of the CYLD USP domain, revealing a distinctive architecture that provides molecular insights into its specificity toward Lys63-linked polyubiquitin. We identify regions of the USP domain responsible for this specificity and demonstrate endodeubiquitinase activity toward such chains. Pathogenic truncations of the CYLD C terminus, associated with the hypertrophic skin tumor cylindromatosis, disrupt the USP domain, accounting for loss of CYLD catalytic activity. A small zinc-binding B box domain, similar in structure to other crossbrace Zn-binding folds--including the RING domain found in E3 ubiquitin ligases--is inserted within the globular core of the USP domain. Biochemical and functional characterization of the B box suggests a role as a protein-interaction module that contributes to determining the subcellular localization of CYLD.
- Section of Structural Biology, Institute of Cancer Research, Chester Beatty Laboratories, 237 Fulham Road, London SW3 6JB, UK.
Organizational Affiliation: