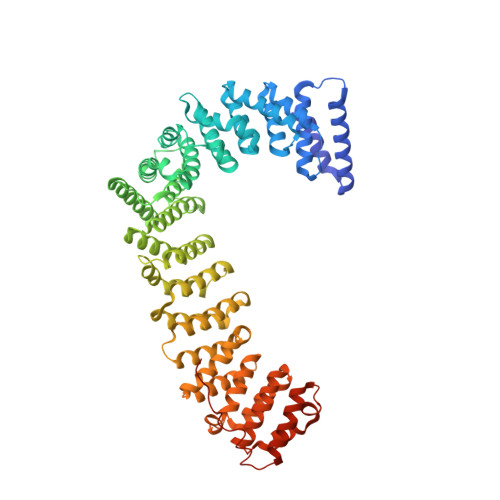
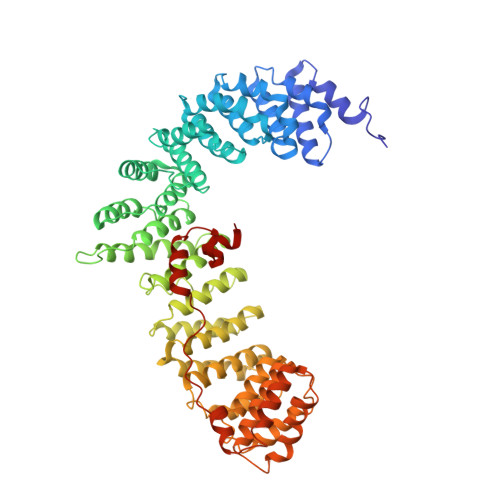
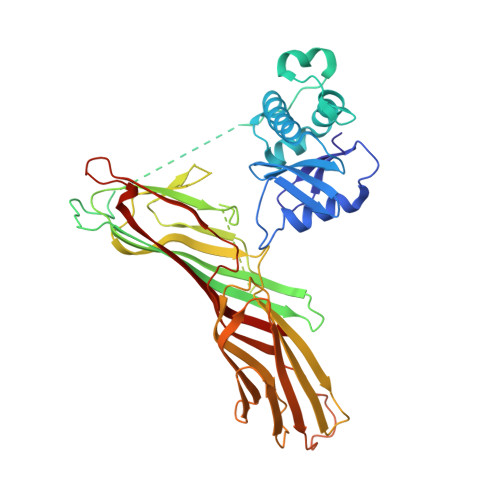
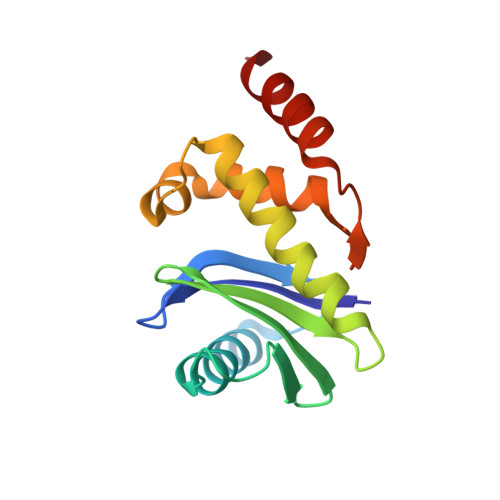
Molecular Architecture and Functional Model of the Endocytic Ap2 Complex
Collins, B.M., Mccoy, A.J., Kent, H.M., Evans, P.R., Owen, D.J.(2002) Cell 109: 523
- PubMed: 12086608
- DOI: https://doi.org/10.1016/s0092-8674(02)00735-3
- Primary Citation of Related Structures:
2VGL - PubMed Abstract:
AP2 is the best-characterized member of the family of heterotetrameric clathrin adaptor complexes that play pivotal roles in many vesicle trafficking pathways within the cell. AP2 functions in clathrin-mediated endocytosis, the process whereby cargo enters the endosomal system from the plasma membrane. We describe the structure of the 200 kDa AP2 "core" (alpha trunk, beta2 trunk, mu2, and sigma2) complexed with the polyphosphatidylinositol headgroup mimic inositolhexakisphosphate at 2.6 A resolution. Two potential polyphosphatidylinositide binding sites are observed, one on alpha and one on mu2. The binding site for Yxxphi endocytic motifs is buried, indicating that a conformational change, probably triggered by phosphorylation in the disordered mu2 linker, is necessary to allow Yxxphi motif binding. A model for AP2 recruitment and activation is proposed.
- Cambridge Institute for Medical Research, University of Cambridge, Department of Clinical Biochemistry, Wellcome Trust/MRC Building, Hills Road, United Kingdom.
Organizational Affiliation: