Molecular Mechanism of Sequence-Directed DNA Loading and Translocation by Ftsk.
Lowe, J., Ellonen, A., Allen, M.D., Atkinson, C., Sherratt, D.J., Grainge, I.(2008) Mol Cell 31: 498
- PubMed: 18722176
- DOI: https://doi.org/10.1016/j.molcel.2008.05.027
- Primary Citation of Related Structures:
2VE8, 2VE9 - PubMed Abstract:
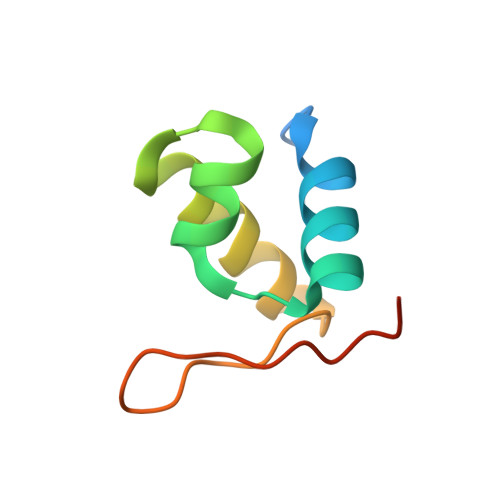
Dimeric circular chromosomes, formed by recombination between monomer sisters, cannot be segregated to daughter cells at cell division. XerCD site-specific recombination at the Escherichia coli dif site converts these dimers to monomers in a reaction that requires the DNA translocase FtsK. Short DNA sequences, KOPS (GGGNAGGG), which are polarized toward dif in the chromosome, direct FtsK translocation. FtsK interacts with KOPS through a C-terminal winged helix domain gamma. The crystal structure of three FtsKgamma domains bound to 8 bp KOPS DNA demonstrates how three gamma domains recognize KOPS. Using covalently linked dimers of FtsK, we infer that three gamma domains per hexamer are sufficient to recognize KOPS and load FtsK and subsequently activate recombination at dif. During translocation, FtsK fails to recognize an inverted KOPS sequence. Therefore, we propose that KOPS act solely as a loading site for FtsK, resulting in a unidirectionally oriented hexameric motor upon DNA.
- MRC Laboratory of Molecular Biology, Hills Road, Cambridge CB2 0QH, UK. Electronic address: jyl@mrc-lmb.cam.ac.uk.
Organizational Affiliation: