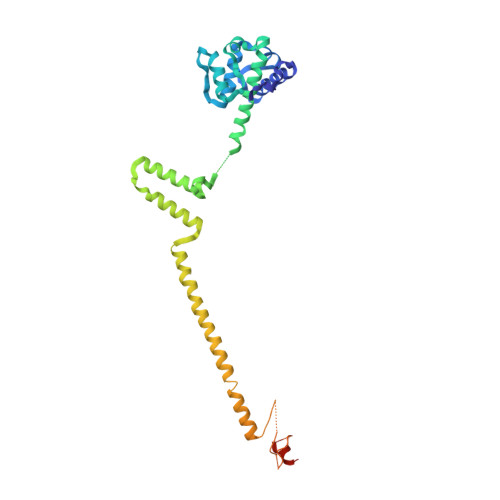
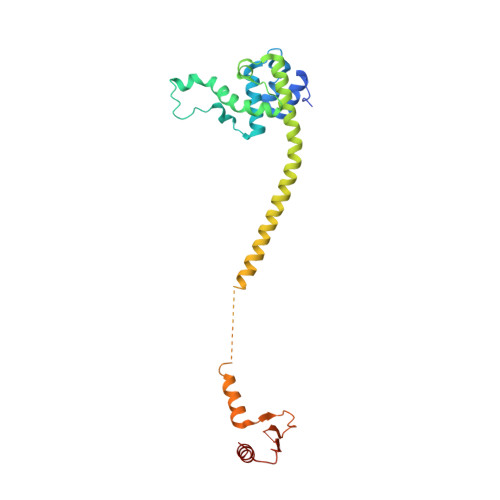
Implications for Kinetochore-Microtubule Attachment from the Structure of an Engineered Ndc80 Complex
Ciferri, C., Pasqualato, S., Screpanti, E., Varetti, G., Santaguida, S., Dos Reis, G., Maiolica, A., Polka, J., De Luca, J.G., De Wulf, P., Salek, M., Rappsilber, J., Moores, C.A., Salmon, E.D., Musacchio, A.(2008) Cell 133: 427
- PubMed: 18455984
- DOI: https://doi.org/10.1016/j.cell.2008.03.020
- Primary Citation of Related Structures:
2VE7 - PubMed Abstract:
Kinetochores are proteinaceous assemblies that mediate the interaction of chromosomes with the mitotic spindle. The 180 kDa Ndc80 complex is a direct point of contact between kinetochores and microtubules. Its four subunits contain coiled coils and form an elongated rod structure with functional globular domains at either end. We crystallized an engineered "bonsai" Ndc80 complex containing a shortened rod domain but retaining the globular domains required for kinetochore localization and microtubule binding. The structure reveals a microtubule-binding interface containing a pair of tightly interacting calponin-homology (CH) domains with a previously unknown arrangement. The interaction with microtubules is cooperative and predominantly electrostatic. It involves positive charges in the CH domains and in the N-terminal tail of the Ndc80 subunit and negative charges in tubulin C-terminal tails and is regulated by the Aurora B kinase. We discuss our results with reference to current models of kinetochore-microtubule attachment and centromere organization.
- Department of Experimental Oncology, European Institute of Oncology, Via Adamello 16, I 20139 Milan, Italy.
Organizational Affiliation: