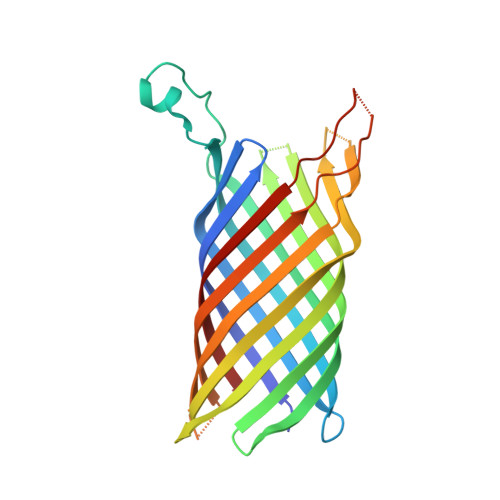
In meso crystal structure and docking simulations suggest an alternative proteoglycan binding site in the OpcA outer membrane adhesin.
Cherezov, V., Liu, W., Derrick, J.P., Luan, B., Aksimentiev, A., Katritch, V., Caffrey, M.(2008) Proteins 71: 24-34
- PubMed: 18076035
- DOI: https://doi.org/10.1002/prot.21841
- Primary Citation of Related Structures:
2VDF - PubMed Abstract:
OpcA is an integral outer membrane adhesin protein from Neisseria meningitidis, the causative agent of meningococcal meningitis and septicemia. It binds to sialic acid (SA)-containing polysaccharides on the surface of epithelial cells. The crystal structure of OpcA showed that the protein adopts a 10-stranded beta-barrel structure, with five extensive loop regions on the extracellular side of the membrane. These form a crevice structure, lined with basic residues, which was hypothesized to act as the binding site for polysaccharide ligands. In the current study, a distinctly different OpcA structure has been obtained using crystals grown from a lipidic mesophase. Comparison of the two structures shows that the largest loop (L2), which closes over the end of the beta-barrel in the original crystal form, adopts a much more extended structure by reaching outward and away from the protein. The difference in conformation may be attributable to the absence of zinc ions from the crystallization conditions for the in meso crystal form: in the original structure, two zinc ions were bound to the external loops. Molecular dynamics (MD) simulations performed on the two OpcA models in a lipid bilayer environment demonstrated pronounced loop mobility. These observations support the view that the loop regions of OpcA are capable of a high degree of conformational flexibility. The original binding site for polysaccharide is not present in the in meso crystal form, and is disrupted during MD simulations. Docking analysis suggests a putative alternative location for the SA ligand in the new crystal form of OpcA.
- Department of Chemistry, The Ohio State University, Columbus, Ohio 43210, USA.
Organizational Affiliation: