Regulation of the Xylan-Degrading Apparatus of Cellvibrio Japonicus by a Novel Two-Component System.
Emami, K., Topakas, E., Nagy, T., Henshaw, J., Jackson, K.A., Nelson, K.E., Mongodin, E.F., Murray, J.W., Lewis, R.J., Gilbert, H.J.(2009) J Biological Chem 284: 1086
- PubMed: 18922794
- DOI: https://doi.org/10.1074/jbc.M805100200
- Primary Citation of Related Structures:
2VA0 - PubMed Abstract:
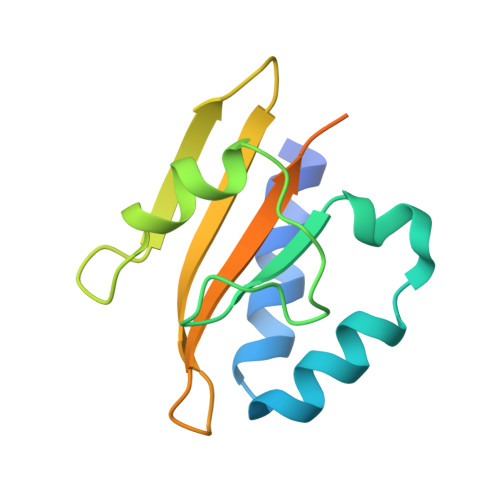
The microbial degradation of lignocellulose biomass is not only an important biological process but is of increasing industrial significance in the bioenergy sector. The mechanism by which the plant cell wall, an insoluble composite structure, activates the extensive repertoire of microbial hydrolytic enzymes required to catalyze its degradation is poorly understood. Here we have used a transposon mutagenesis strategy to identify a genetic locus, consisting of two genes that modulate the expression of xylan side chain-degrading enzymes in the saprophytic bacterium Cellvibrio japonicus. Significantly, the locus encodes a two-component signaling system, designated AbfS (sensor histidine kinase) and AbfR (response regulator). The AbfR/S two-component system is required to activate the expression of the suite of enzymes that remove the numerous side chains from xylan, but not the xylanases that hydrolyze the beta1,4-linked xylose polymeric backbone of this polysaccharide. Studies on the recombinant sensor domain of AbfS (AbfS(SD)) showed that it bound to decorated xylans and arabinoxylo-oligosaccharides, but not to undecorated xylo-oligosaccharides or other plant structural polysaccharides/oligosaccharides. The crystal structure of AbfS(SD) was determined to a resolution of 2.6A(.) The overall fold of AbfS(SD) is that of a classical Per Arndt Sim domain with a central antiparallel four-stranded beta-sheet flanked by alpha-helices. Our data expand the number of molecules known to bind to the sensor domain of two-component histidine kinases to include complex carbohydrates. The biological rationale for a regulatory system that induces enzymes that remove the side chains of xylan, but not the hydrolases that cleave the backbone of the polysaccharide, is discussed.
- Institute for Cell and Molecular Biosciences, Newcastle University, The Medical School, Framlington Place, Newcastle-upon-Tyne, NE2 4HH, United Kingdom.
Organizational Affiliation: