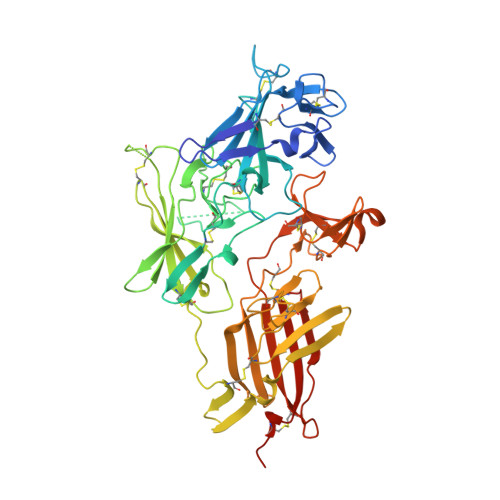
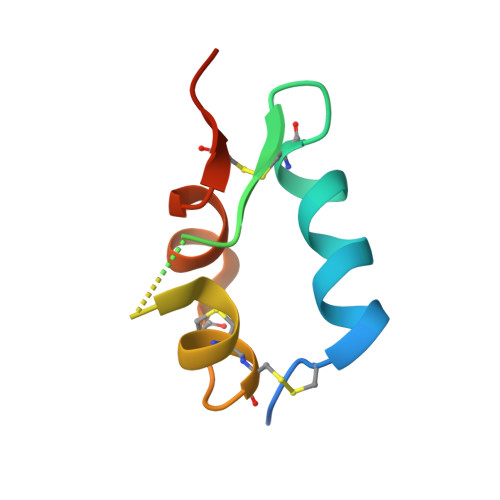
Structure and Functional Analysis of the Igf-II/Igf2R Interaction
Brown, J., Delaine, C., Zaccheo, O.J., Siebold, C., Gilbert, R.J., Van Boxel, G., Denley, A., Wallace, J.C., Hassan, A.B., Forbes, B.E., Jones, E.Y.(2008) EMBO J 27: 265
- PubMed: 18046459
- DOI: https://doi.org/10.1038/sj.emboj.7601938
- Primary Citation of Related Structures:
2V5N, 2V5O, 2V5P - PubMed Abstract:
Embryonic development and normal growth require exquisite control of insulin-like growth factors (IGFs). In mammals the extracellular region of the cation-independent mannose-6-phosphate receptor has gained an IGF-II-binding function and is termed type II IGF receptor (IGF2R). IGF2R sequesters IGF-II; imbalances occur in cancers and IGF2R is implicated in tumour suppression. We report crystal structures of IGF2R domains 11-12, 11-12-13-14 and domains 11-12-13/IGF-II complex. A distinctive juxtaposition of these domains provides the IGF-II-binding unit, with domain 11 directly interacting with IGF-II and domain 13 modulating binding site flexibility. Our complex shows that Phe19 and Leu53 of IGF-II lock into a hydrophobic pocket unique to domain 11 of mammalian IGF2Rs. Mutagenesis analyses confirm this IGF-II 'binding-hotspot', revealing that IGF-binding proteins and IGF2R have converged on the same high-affinity site.
- Cancer Research UK Receptor Structure Research Group, Division of Structural Biology, Wellcome Trust Centre for Human Genetics, University of Oxford, Headington, Oxford, UK.
Organizational Affiliation: