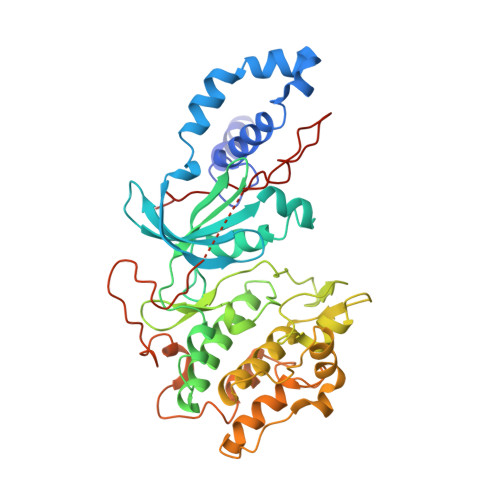
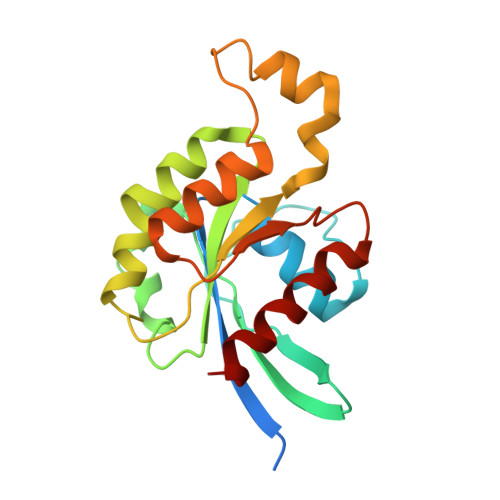
Mechanism of Multi-Site Phosphorylation from a Rock-I:Rhoe Complex Structure.
Komander, D., Garg, R., Wan, P.T.C., Ridley, A.J., Barford, D.(2008) EMBO J 27: 3175
- PubMed: 18946488
- DOI: https://doi.org/10.1038/emboj.2008.226
- Primary Citation of Related Structures:
2V55 - PubMed Abstract:
The ROCK-I serine/threonine protein kinase mediates the effects of RhoA to promote the formation of actin stress fibres and integrin-based focal adhesions. ROCK-I phosphorylates the unconventional G-protein RhoE on multiple N- and C-terminal sites. These phosphorylation events stabilise RhoE, which functions to antagonise RhoA-induced stress fibre assembly. Here, we provide a molecular explanation for multi-site phosphorylation of RhoE from the crystal structure of RhoE in complex with the ROCK-I kinase domain. RhoE interacts with the C-lobe alphaG helix of ROCK-I by means of a novel binding site remote from its effector region, positioning its N and C termini proximal to the ROCK-I catalytic site. Disruption of the ROCK-I:RhoE interface abolishes RhoE phosphorylation, but has no effect on the ability of RhoE to disassemble stress fibres. In contrast, mutation of the RhoE effector region attenuates RhoE-mediated disruption of the actin cytoskeleton, indicating that RhoE exerts its inhibitory effects on ROCK-I through protein(s) binding to its effector region. We propose that ROCK-I phosphorylation of RhoE forms part of a feedback loop to regulate RhoA signalling.
- Section of Structural Biology, The Institute of Cancer Research, Chester Beatty Laboratories, London, UK.
Organizational Affiliation: