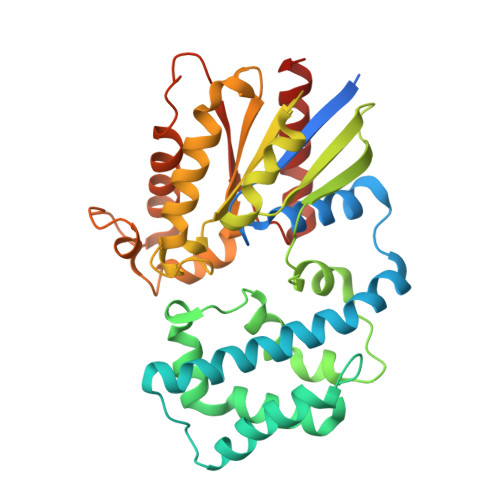
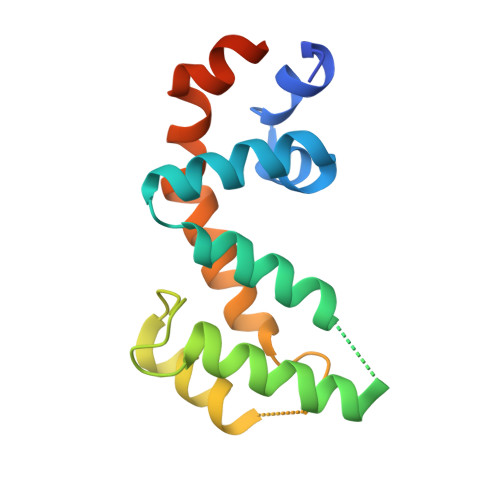
Structural Determinants of G-Protein Alpha Subunit Selectivity by Regulator of G-Protein Signaling 2(Rgs2).
Kimple, A.J., Soundararajan, M., Hutsell, S.Q., Roos, A.K., Urban, D.J., Setola, V., Temple, B.R., Roth, B.L., Knapp, S., Willard, F.S., Siderovski, D.P.(2009) J Biological Chem 284: 19402
- PubMed: 19478087
- DOI: https://doi.org/10.1074/jbc.M109.024711
- Primary Citation of Related Structures:
2V4Z - PubMed Abstract:
"Regulator of G-protein signaling" (RGS) proteins facilitate the termination of G protein-coupled receptor (GPCR) signaling via their ability to increase the intrinsic GTP hydrolysis rate of Galpha subunits (known as GTPase-accelerating protein or "GAP" activity). RGS2 is unique in its in vitro potency and selectivity as a GAP for Galpha(q) subunits. As many vasoconstrictive hormones signal via G(q) heterotrimer-coupled receptors, it is perhaps not surprising that RGS2-deficient mice exhibit constitutive hypertension. However, to date the particular structural features within RGS2 determining its selectivity for Galpha(q) over Galpha(i/o) substrates have not been completely characterized. Here, we examine a trio of point mutations to RGS2 that elicits Galpha(i)-directed binding and GAP activities without perturbing its association with Galpha(q). Using x-ray crystallography, we determined a model of the triple mutant RGS2 in complex with a transition state mimetic form of Galpha(i) at 2.8-A resolution. Structural comparison with unliganded, wild type RGS2 and of other RGS domain/Galpha complexes highlighted the roles of these residues in wild type RGS2 that weaken Galpha(i) subunit association. Moreover, these three amino acids are seen to be evolutionarily conserved among organisms with modern cardiovascular systems, suggesting that RGS2 arose from the R4-subfamily of RGS proteins to have specialized activity as a potent and selective Galpha(q) GAP that modulates cardiovascular function.
- Departments of Pharmacology, University of North Carolina, Chapel Hill, North Carolina 27599, USA.
Organizational Affiliation: