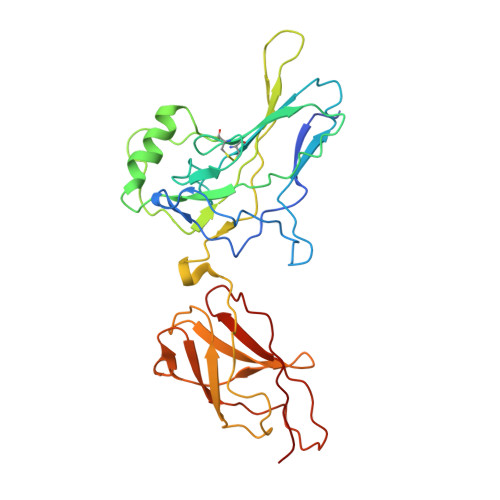
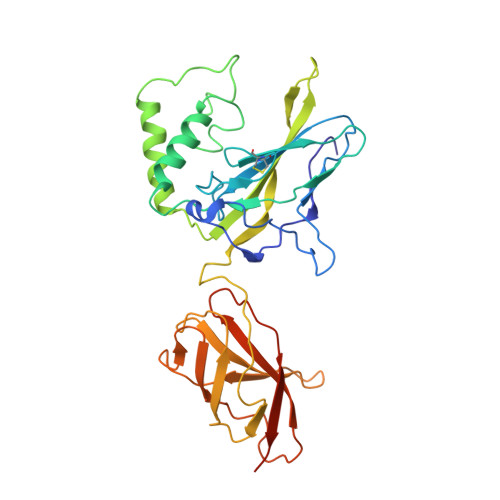
X-Ray Structure of a NF-kappaB p50/Relb/DNA Complex Reveals Assembly of Multiple Dimers on Tandem kappaB Sites.
Moorthy, A.K., Huang, D.B., Wang, V.Y., Vu, D., Ghosh, G.(2007) J Mol Biology 373: 723
- PubMed: 17869269
- DOI: https://doi.org/10.1016/j.jmb.2007.08.039
- Primary Citation of Related Structures:
2V2T - PubMed Abstract:
We describe here the X-ray crystal structure of NF-kappaB p50/RelB heterodimer bound to a kappaB DNA. Although the global modes of subunit association and kappaB DNA recognition are similar to other NF-kappaB/DNA complexes, this complex reveals distinctive features not observed for non-RelB complexes. For example, Lys274 of RelB is removed from the protein-DNA interface whereas the corresponding residues in all other subunits make base-specific contacts. This mode of binding suggests that RelB may allow the recognition of more diverse kappaB sequences. Complementary surfaces on RelB and p50, as revealed by the crystal contacts, are highly suggestive of assembly of multiple p50/RelB heterodimers on tandem kappaB sites in solution. Consistent with this model our in vitro binding experiments reveal optimal assembly of two wild-type p50/RelB heterodimers on tandem HIV kappaB DNA with 2 bp spacing but not by a mutant heterodimer where one of the RelB packing surface is altered. We suggest that multiple NF-kappaB dimers assemble at diverse kappaB promoters through direct interactions utilizing unique protein-protein interaction surfaces.
- Department of Chemistry and Biochemistry, University of California, San Diego, 9500 Gilman Drive, La Jolla, CA 92093, USA.
Organizational Affiliation: