Determination of Three-Dimensional Structure and Residues of the Novel Tumor Suppressor Aimp3/P18 Required for the Interaction with Atm.
Kim, K.J., Park, M.C., Choi, S.J., Oh, Y.S., Choi, E.C., Cho, H.J., Kim, M.H., Kim, S.H., Kim, D.W., Kim, S., Kang, B.S.(2008) J Biological Chem 283: 14032
- PubMed: 18343821
- DOI: https://doi.org/10.1074/jbc.M800859200
- Primary Citation of Related Structures:
2UZ8 - PubMed Abstract:
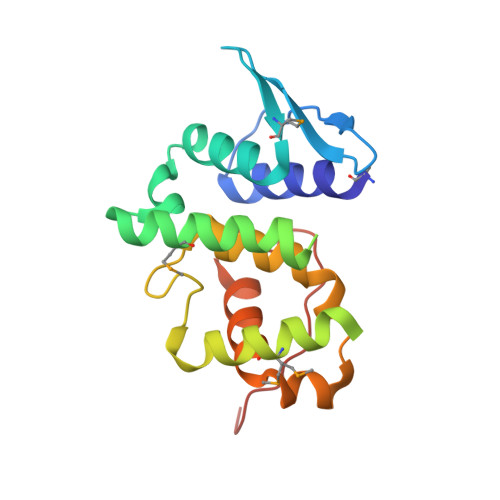
Although AIMP3/p18 is normally associated with the multi-tRNA synthetase complex via its specific interaction with methionyl-tRNA synthetase, it also works as a tumor suppressor by interacting with ATM, the upstream kinase of p53. To understand the molecular interactions of AIMP3 and the mechanisms involved, we determined the crystal structure of AIMP3 at 2.0-angstroms resolution and identified its potential sites of interaction with ATM. AIMP3 contains two distinct domains linked by a 7-amino acid (Lys57-Ser63) peptide, which contains a 3(10) helix. The 56-amino acid N-terminal domain consists of two helices into which three antiparallel beta strands are inserted, and the 111-amino acid C-terminal domain contains a bundle of five helices (Thr64-Tyr152) followed by a coiled region (Pro153-Leu169). Structural analyses revealed homologous proteins such as yeast glutamyl-tRNA synthetase, Arc1p, EF1Bgamma, and glutathione S-transferase and suggested two potential molecular binding sites. Moreover, mutations at the C-terminal putative binding site abolished the interaction between AIMP3 and ATM and the ability of AIMP3 to activate p53. Thus, this work identified the two potential molecular interaction sites of AIMP3 and determined the residues critical for its tumor-suppressive activity through the interaction with ATM.
- Pohang Accelerator Laboratory, Pohang University of Science and Technology, Pohang 790-784, Korea.
Organizational Affiliation: