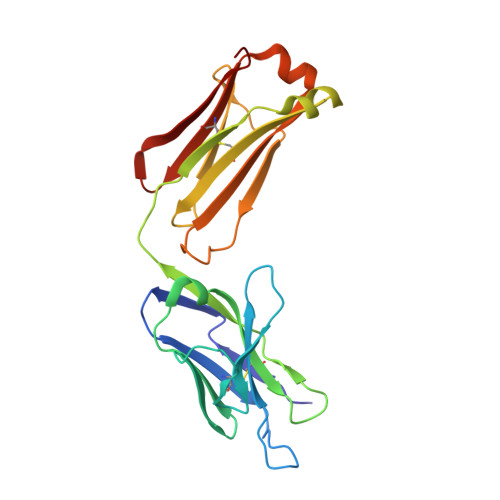
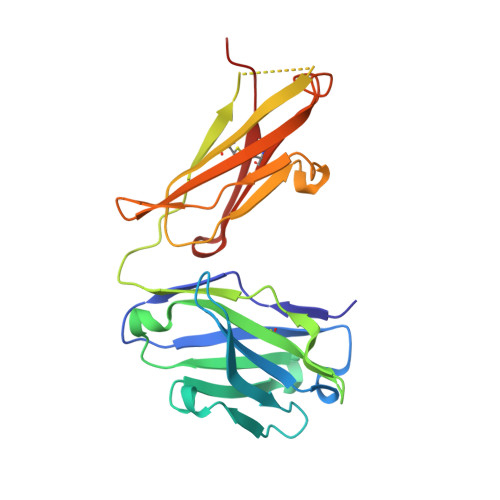
Crystal Structure of a Monoclonal Antibody Directed Against an Antigenic Determinant Common to Ogawa and Inaba Serotypes of Vibrio Cholerae O1.
Ahmed, F., Andre-Leroux, G., Haouz, A., Boutonnier, A., Delepierre, M., Qadri, F., Nato, F., Fournier, J.M., Alzari, P.M.(2008) Proteins 70: 284
- PubMed: 17876834
- DOI: https://doi.org/10.1002/prot.21609
- Primary Citation of Related Structures:
2UYL - Institut Pasteur and CNRS URA 2185, 25/28 rue du Dr. Roux, Paris, France.
Organizational Affiliation: