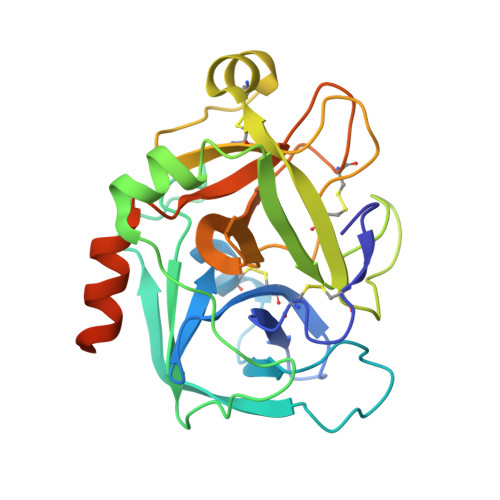
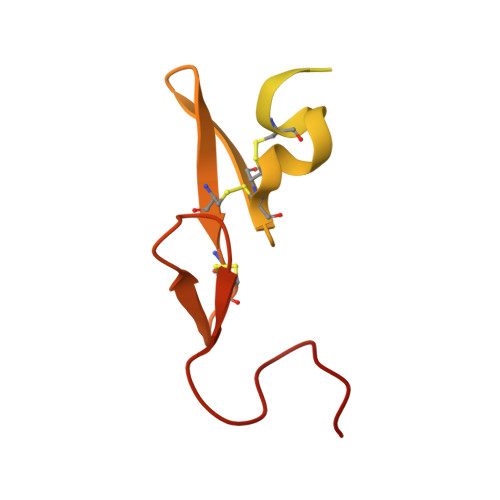
Selective and Dual Action Orally Active Inhibitors of Thrombin and Factor Xa.
Young, R.J., Brown, D., Burns-Kurtis, C.L., Chan, C., Convery, M.A., Hubbard, J.A., Kelly, H.A., Pateman, A.J., Patikis, A., Senger, S., Shah, G.P., Toomey, J.R., Watson, N.S., Zhou, P.(2007) Bioorg Med Chem Lett 17: 2927
- PubMed: 17420122
- DOI: https://doi.org/10.1016/j.bmcl.2007.03.080
- Primary Citation of Related Structures:
2UWL, 2UWO, 2UWP - PubMed Abstract:
The synthetic entry to new classes of dual fXa/thrombin and selective thrombin inhibitors with significant oral bioavailability is described. This was achieved through minor modifications to the sulfonamide group in our potent and selective fXa inhibitor (E)-2-(5-chlorothien-2-yl)-N-{(3S)-1-[(1S)-1-methyl-2-(morpholin-4-yl)-2-oxoethyl]-2-oxopyrrolidin-3-yl}ethenesulfonamide and these observed activity changes have been rationalised using structural studies.
- GlaxoSmithKline, Medicines Research Centre, Gunnels Wood Road, Stevenage, Hertfordshire SG1 2NY, UK. Rob.J.Young@gsk.com
Organizational Affiliation: