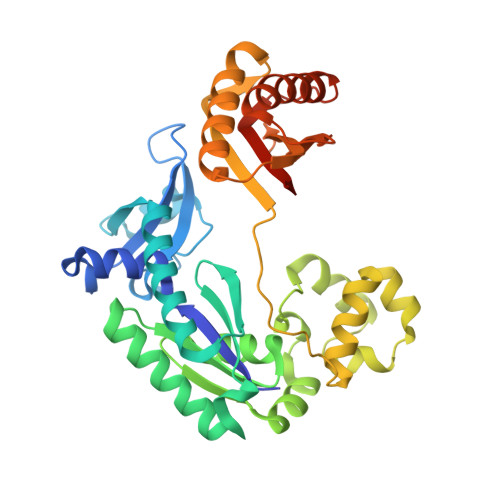
Hydrogen Bonding of 7,8-Dihydro-8-Oxodeoxyguanosine with a Charged Residue in the Little Finger Domain Determines Miscoding Events in Sulfolobus Solfataricus DNA Polymerase Dpo4.
Eoff, R.L., Irimia, A., Angel, K.C., Egli, M., Guengerich, F.P.(2007) J Biological Chem 282: 19831
- PubMed: 17468100
- DOI: https://doi.org/10.1074/jbc.M702290200
- Primary Citation of Related Structures:
2UVR, 2UVU, 2UVV, 2UVW - PubMed Abstract:
Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4) has been shown to catalyze bypass of 7,8-dihydro-8-oxodeoxyguanosine (8-oxoG) in a highly efficient and relatively accurate manner. Crystal structures have revealed a potential role for Arg(332) in stabilizing the anti conformation of the 8-oxoG template base by means of a hydrogen bond or ion-dipole pair, which results in an increased enzymatic efficiency for dCTP insertion and makes formation of a Hoogsteen pair between 8-oxoG and dATP less favorable. Site-directed mutagenesis was used to replace Arg(332) with Ala, Glu, Leu, or His in order to probe the importance of Arg(332) in accurate and efficient bypass of 8-oxoG. The double mutant Ala(331)Ala(332) was also prepared to address the contribution of Arg(331). Transientstate kinetic results suggest that Glu(332) retains fidelity against bypass of 8-oxoG that is similar to wild type Dpo4, a result that was confirmed by tandem mass spectrometric analysis of full-length extension products. A crystal structure of the Dpo4 Glu(332) mutant and 8-oxoG:C pair revealed water-mediated hydrogen bonds between Glu(332) and the O-8 atom of 8-oxoG. The space normally occupied by Arg(332) side chain is empty in the crystal structures of the Ala(332) mutant. Two other crystal structures show that a Hoogsteen base pair is formed between 8-oxoG and A in the active site of both Glu(332) and Ala(332) mutants. These results support the view that a bond between Arg(332) and 8-oxoG plays a role in determining the fidelity and efficiency of Dpo4-catalyzed bypass of the lesion.
- Department of Biochemistry and Center in Molecular Toxicology, Vanderbilt University School of Medicine, Nashville, Tennessee 37232-0146, USA.
Organizational Affiliation: