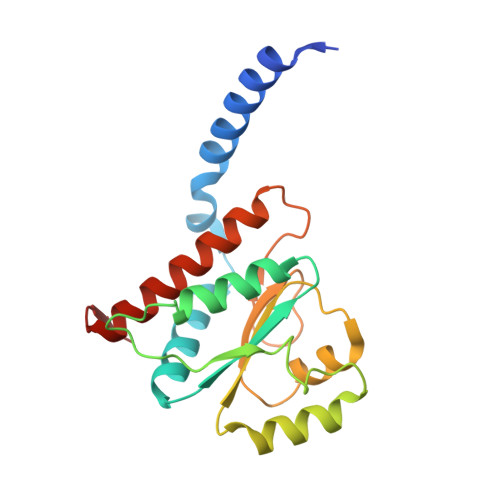
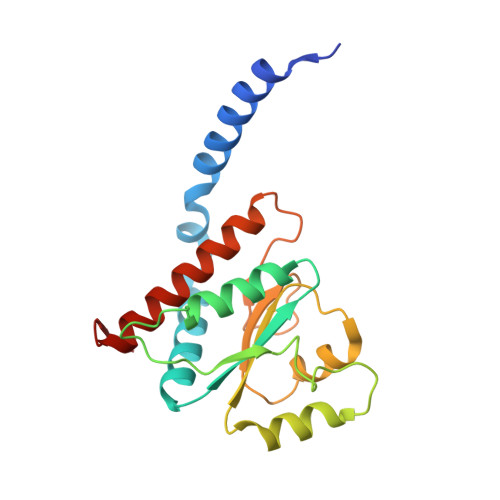
Structural similarity between the DnaA-binding proteins HobA (HP1230) from Helicobacter pylori and DiaA from Escherichia coli.
Natrajan, G., Hall, D.R., Thompson, A.C., Gutsche, I., Terradot, L.(2007) Mol Microbiol 65: 995-1005
- PubMed: 17683397
- DOI: https://doi.org/10.1111/j.1365-2958.2007.05843.x
- Primary Citation of Related Structures:
2UVP - PubMed Abstract:
In prokaryotes, DNA replication is initiated by the binding of DnaA to the oriC region of the chromosome to load the primosome machinery and start a new replication round. Several proteins control these events in Escherichia coli to ensure that replication is precisely timed during the cell cycle. Here, we report the crystal structure of HobA (HP1230) at 1.7 A, a recently discovered protein that specifically interacts with DnaA protein from Helicobacter pylori (HpDnaA). We found that the closest structural homologue of HobA is a sugar isomerase (SIS) domain containing protein, the phosphoheptose isomerase from Pseudomonas aeruginosa. Remarkably, SIS proteins share strong sequence homology with DiaA from E. coli; yet, HobA and DiaA share no sequence homology. Thus, by solving the structure of HobA, we unexpectedly discovered that HobA is a H. pylori structural homologue of DiaA. By comparing the structure of HobA to a homology model of DiaA, we identified conserved, surface-accessible residues that could be involved in protein-protein interaction. Finally, we show that HobA specifically interacts with the N-terminal part of HpDnaA. The structural homology between DiaA and HobA strongly supports their involvement in the replication process and these proteins could define a new structural family of replication regulators in bacteria.
- Macromolecular Crystallography Group, European Synchrotron Radiation Facility, B.P. 220, 6 rue Jules Horowitz, F-38043 Grenoble Cedex, France.
Organizational Affiliation: