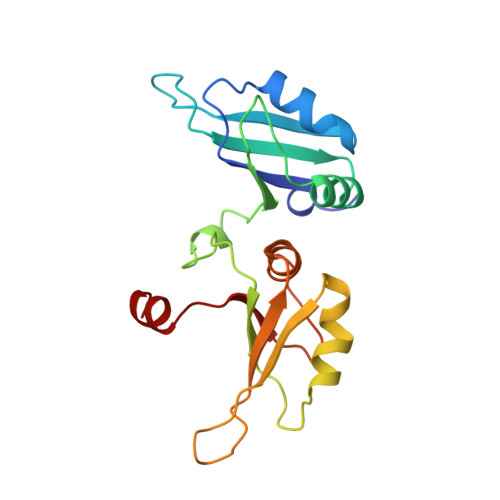
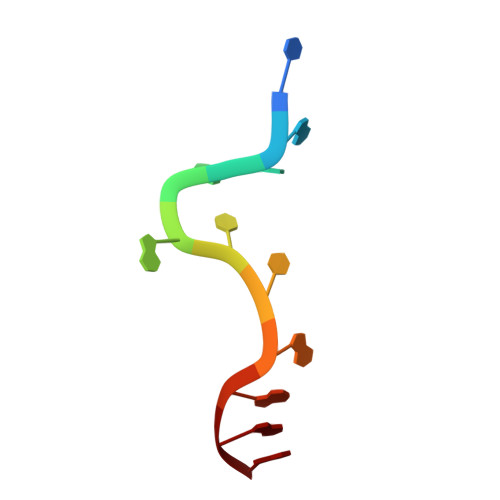
Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA.
Ding, J., Hayashi, M.K., Zhang, Y., Manche, L., Krainer, A.R., Xu, R.M.(1999) Genes Dev 13: 1102-1115
- PubMed: 10323862
- DOI: https://doi.org/10.1101/gad.13.9.1102
- Primary Citation of Related Structures:
2UP1 - PubMed Abstract:
Human hnRNP A1 is a versatile single-stranded nucleic acid-binding protein that functions in various aspects of mRNA maturation and in telomere length regulation. The crystal structure of UP1, the amino-terminal domain of human hnRNP A1 containing two RNA-recognition motifs (RRMs), bound to a 12-nucleotide single-stranded telomeric DNA has been determined at 2.1 A resolution. The structure of the complex reveals the basis for sequence-specific recognition of the single-stranded overhangs of human telomeres by hnRNP A1. It also provides insights into the basis for high-affinity binding of hnRNP A1 to certain RNA sequences, and for nucleic acid binding and functional synergy between the RRMs. In the crystal structure, a UP1 dimer binds to two strands of DNA, and each strand contacts RRM1 of one monomer and RRM2 of the other. The two DNA strands are antiparallel, and regions of the protein flanking each RRM make important contacts with DNA. The extensive protein-protein interface seen in the crystal structure of the protein-DNA complex and the evolutionary conservation of the interface residues suggest the importance of specific protein-protein interactions for the sequence-specific recognition of single-stranded nucleic acids. Models for regular packaging of telomere 3' overhangs and for juxtaposition of alternative 5' splice sites are proposed.
- W.M. Keck Structural Biology Laboratory, Cold Spring Harbor, New York 11724, USA.
Organizational Affiliation: