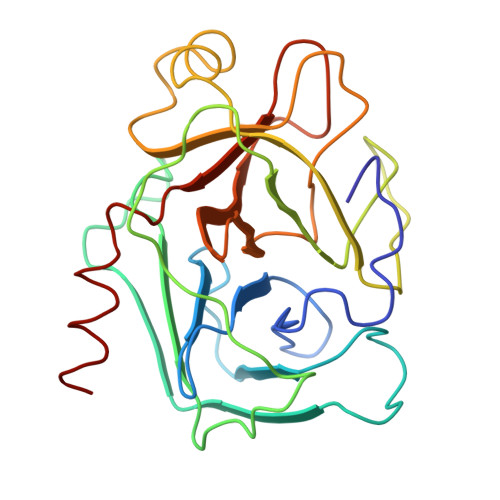
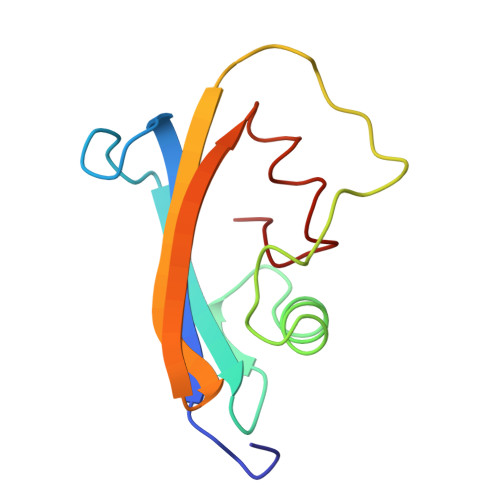
Crystal structure of an engineered subtilisin inhibitor complexed with bovine trypsin.
Takeuchi, Y., Nonaka, T., Nakamura, K.T., Kojima, S., Miura, K., Mitsui, Y.(1992) Proc Natl Acad Sci U S A 89: 4407-4411
- PubMed: 1584773
- DOI: https://doi.org/10.1073/pnas.89.10.4407
- Primary Citation of Related Structures:
2TLD - PubMed Abstract:
Proteinase specificity of a proteinaceous inhibitor of subtilisin (SSI; Streptomyces subtilisin inhibitor) can be altered so as to strongly inhibit trypsin simply by replacing P1 methionine with lysine (with or without concomitant change of the P4 residue) through site-directed mutagenesis. Now the crystal structure of one such engineered SSI (P1 methionine converted to lysine and P4 methionine converted to glycine) complexed with bovine trypsin has been solved at 2.6 A resolution and refined to a crystallographic R factor of 0.173. Comparing this structure with the previously established structure of the native SSI complexed with subtilisin BPN', it was found that (i) P1 lysine of the mutant SSI is accommodated in the S1 pocket of trypsin as usual, and (ii) upon complex formation, considerable conformation change occurs to the reactive site loop of the mutant SSI. Thus, in this case, flexibility of the reactive site loop seems important for successfully changing the proteinase specificity through mere replacement of the P1 residue.
- Pharmaceutical Research Center, Meiji Seika Kaisha, Ltd., Yokohama, Japan.
Organizational Affiliation: