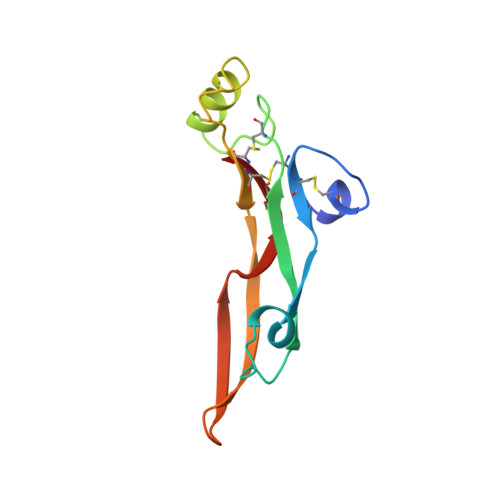
Crystal structure of transforming growth factor-beta 2: an unusual fold for the superfamily.
Daopin, S., Piez, K.A., Ogawa, Y., Davies, D.R.(1992) Science 257: 369-373
- PubMed: 1631557
- DOI: https://doi.org/10.1126/science.1631557
- Primary Citation of Related Structures:
2TGI - PubMed Abstract:
The transforming growth factors-beta (TGF-beta 1 through -beta 5) are a family of homodimeric cytokines that regulate proliferation and function in many cell types. Family members have 66 to 80% sequence identity and nine strictly conserved cysteines. A crystal structure of a member of this family, TGF-beta 2, has been determined at 2.1 angstrom (A) resolution and refined to an R factor of 0.172. The monomer lacks a well-defined hydrophobic core and displays an unusual elongated nonglobular fold with dimensions of approximately 60 A by 20 A by 15 A. Eight cysteines form four intrachain disulfide bonds, which are clustered in a core region forming a network complementary to the network of hydrogen bonds. The dimer is stabilized by the ninth cysteine, which forms an interchain disulfide bond, and by two identical hydrophobic interfaces. Sequence profile analysis of other members of the TGF-beta superfamily, including the activins, inhibins, and several developmental factors, imply that they also adopt the TGF-beta fold.
- Laboratory of Molecular Biology, National Institute of Diabetes, Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD 20892.
Organizational Affiliation: