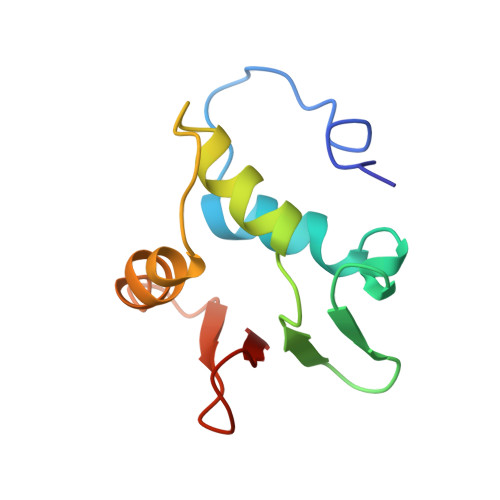
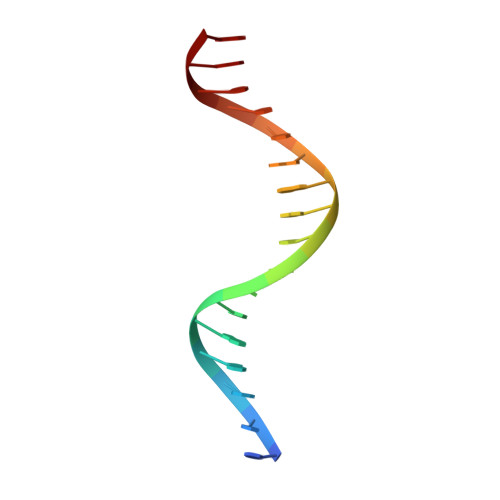
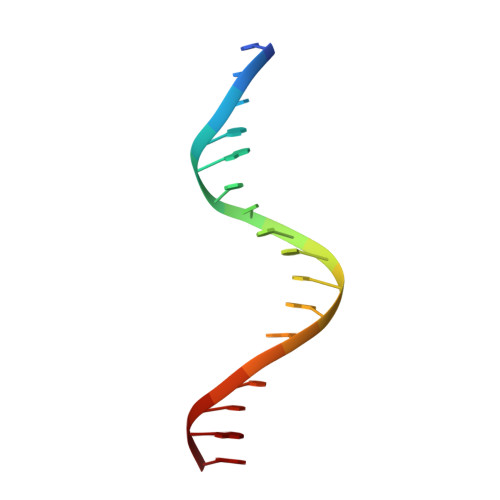
Correction of the NMR structure of the ETS1/DNA complex.
Werner, M.H., Clore, G.M., Fisher, C.L., Fisher, R.J., Trinh, L., Shiloach, J., Gronenborn, A.M.(1997) J Biomol NMR 10: 317-328
- PubMed: 9460239
- DOI: https://doi.org/10.1023/a:1018399711996
- Primary Citation of Related Structures:
2STT, 2STW - PubMed Abstract:
The ETS family of transcription factors consists of a group of proteins that share a highly conserved 85 amino acid DNA-binding domain (DBD). This family recognizes a consensus sequence rich in purine bases with a central GGAA motif. A comparison of the published three-dimensional structures of the DBD/DNA complexes of ETS1 by NMR [Werner et al. (1995) Cell, 83, 761-771] and the related Pu.1 by X-ray crystallography [Kodandapani et al. (1996) Nature, 380, 456-460] reveals an apparent discrepancy in which the protein domains bind with opposite polarity to their target sequences. This surprising and highly unlikely result prompted us to reexamine our NMR structure. Additional NMR experiments now reveal an error in the original interpretation of the spectra defining the orientation of the ETS1-DBD on DNA. It was originally reported that the ETS1-DBD bound to DNA with a bipartite motif involving major groove recognition via a helix-turn-helix element and minor groove recognition via protein side-chain intercalation. The presence of intercalation was deduced on the basis of numerous NOEs between several amino acids in the protein and a resonance at 12.33 ppm originally assigned to a DNA imino proton. New NMR experiments now conclusively demonstrate that this resonance, which is located within the DNA imino proton region of the spectrum, arises from the hydroxyl proton of Tyr86. Realization of this error necessitated reanalysis of the intermolecular NOEs. This revealed that the orientation of the ETS1-DBD in the complex is opposite to that originally reported and that a tryptophan residue does not intercalate into the DNA. The calculation of a new ensemble of structures based on the corrected data indicates that the structure of the ETS1-DBD/DNA complex is indeed similar to the X-ray structure of the Pu.1-DBD/DNA complex.
- Laboratory of Chemical Physics, NIDDKD, National Institutes of Health, Bethesda, MD 20892-0520, USA.
Organizational Affiliation: