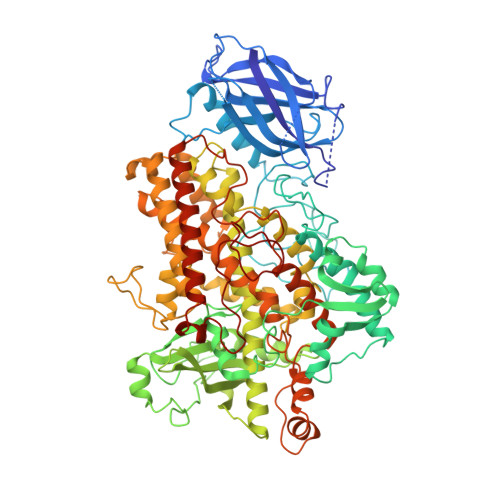
The three-dimensional structure of an arachidonic acid 15-lipoxygenase.
Boyington, J.C., Gaffney, B.J., Amzel, L.M.(1993) Science 260: 1482-1486
- PubMed: 8502991
- DOI: https://doi.org/10.1126/science.8502991
- Primary Citation of Related Structures:
2SBL - PubMed Abstract:
In mammals, the hydroperoxidation of arachidonic acid by lipoxygenases leads to the formation of leukotrienes and lipoxins, compounds that mediate inflammatory responses. Lipoxygenases are dioxygenases that contain a nonheme iron and are present in many animal cells. Soybean lipoxygenase-1 is a single-chain, 839-residue protein closely related to mammalian lipoxygenases. The structure of soybean lipoxygenase-1 solved to 2.6 angstrom resolution shows that the enzyme has two domains: a 146-residue beta barrel and a 693-residue helical bundle. The iron atom is in the center of the larger domain and is coordinated by three histidines and the COO- of the carboxyl terminus. The coordination geometry is nonregular and appears to be a distorted octahedron in which two adjacent positions are not occupied by ligands. Two cavities, in the shapes of a bent cylinder and a frustum, connect the unoccupied positions to the surface of the enzyme. The iron, with two adjacent and unoccupied positions, is poised to interact with the 1,4-diene system of the substrate and with molecular oxygen during catalysis.
- Department of Biophysics and Biophysical Chemistry, Johns Hopkins University School of Medicine, Baltimore, MD 21205.
Organizational Affiliation: