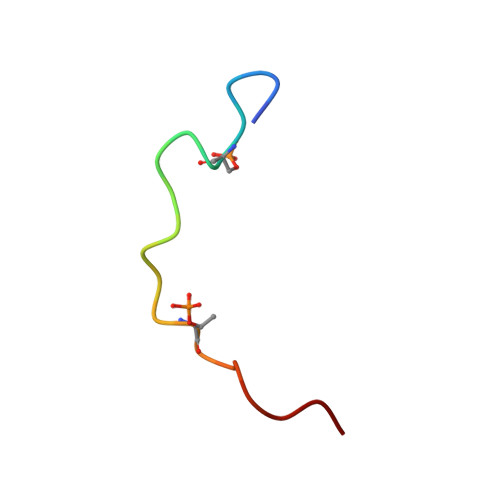
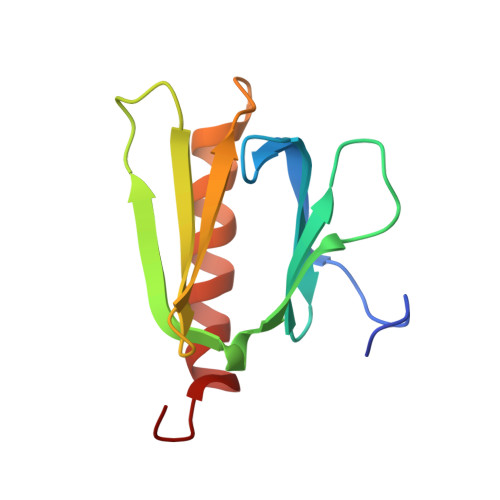
Extended string binding mode of the phosphorylated transactivation domain of tumor suppressor p53.
Okuda, M., Nishimura, Y.(2014) J Am Chem Soc 136: 14143-14152
- PubMed: 25216154
- DOI: https://doi.org/10.1021/ja506351f
- Primary Citation of Related Structures:
2RUK - PubMed Abstract:
The transactivation domain (TAD) of tumor suppressor p53 has homologous subdomains, TAD1 and TAD2. Both are intrinsically disordered in their free states, but all structures of TAD1 and TAD2 bound to their target proteins have demonstrated use of an amphipathic α-helix, suggesting that the binding-coupled helix folding mechanism of TAD1 and TAD2 is essential. Although phosphorylation of TAD is important to switch the function of p53, bound structures of phosphorylated TAD1 and TAD2 have not been determined. Here, we reveal the recognition mechanism of the phosphorylated TAD2 bound to a pleckstrin homology (PH) domain from human TFIIH subunit p62 in an extended string-like conformation. This string-like binding mode of TAD2 seems to be independent of its phosphorylation in spite of enhanced binding activity upon phosphorylation. This is in contrast to the amphipathic helical binding mode of the unphosphorylated TAD2 to the yeast tfb1 PH domain and demonstrates that the p53 TAD2 has much higher conformational malleability than previously appreciated.
- Graduate School of Medical Life Science, Yokohama City University , 1-7-29 Suehiro-cho, Tsurumi-ku, Yokohama 230-0045, Japan.
Organizational Affiliation: