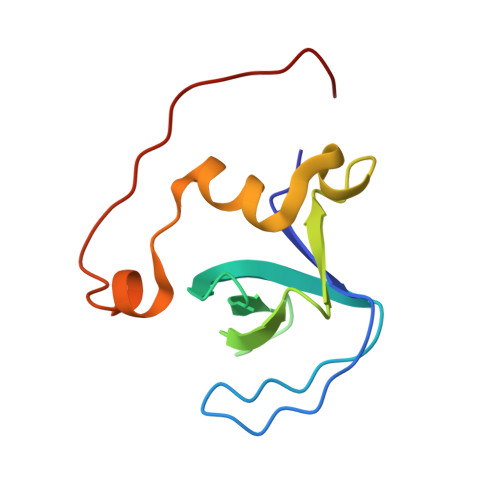
Solution structure of a novel Cdc42-binding module of Bem1 and its interaction with Ste20 and Cdc42
Takaku, T., Ogura, K., Kumeta, H., Yoshida, N., Inagaki, F.(2010) J Biological Chem 285: 19346-19353
- PubMed: 20410294
- DOI: https://doi.org/10.1074/jbc.M110.116749
- Primary Citation of Related Structures:
2RQV, 2RQW - PubMed Abstract:
Bem1 is a scaffold protein essential for the establishment of cell polarity in Saccharomyces cerevisiae. This work reports the solution structure of a Cdc42 binding module of Bem1 comprising the second SH3 domain (SH3b) and its C-terminal flanking region termed Cdc42 interacting (CI). First, the structure of Bem1 SH3b-CI was determined by NMR spectroscopy, which shows that Bem1 SH3b-CI is a structurally and functionally related domain that binds Cdc42. Next, the solution structure of Bem1 SH3b-CI in complex with the proline-rich region of p21-activated kinase Ste20 (Ste20 PRR) was determined. Finally, the interaction surface of Bem1 SH3b-CI with Cdc42 was identified based on chemical shift perturbation studies which reveals that Bem1 SH3b-CI interacts simultaneously with both Ste20 PRR and Cdc42 using the opposite surfaces. Thus, Bem1 can tether Cdc42 and Ste20 in close proximity so that Cdc42 can efficiently interact with Ste20 Cdc42 and Rac interactive binding (CRIB). Based on the present results together with the previous biochemical studies (Lamson, R. E., Winters, M. J., and Pryciak, P. M. (2002) Mol. Cell. Biol. 22, 2939-2951 and Winters, M. J., and Pryciak, P. M. (2005) Mol. Cell. Biol. 25, 2177-2190), a model was suggested that the autoinhibition of Ste20 kinase activity by CRIB is released through the Cdc42-CRIB interaction, which is mediated by Bem1, and Ste20 is subsequently activated, an initial step for the establishment of the cell polarity.
- Laboratory of Structural Biology, Graduate School of Life Science, Hokkaido University, N-12, W-6 Kita-ku, Sapporo 060-0812, Japan.
Organizational Affiliation: