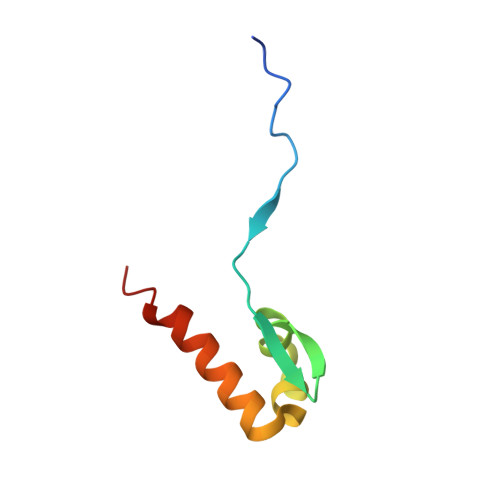
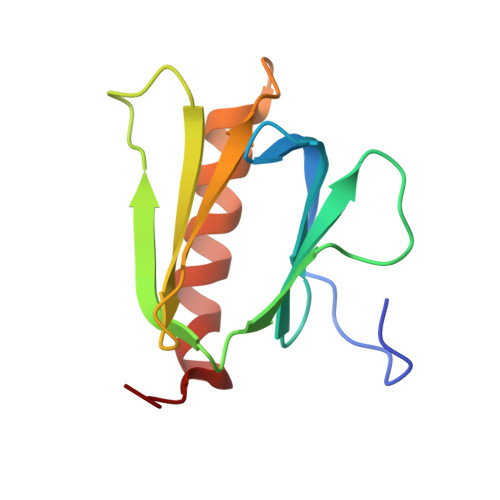
Structural insight into the TFIIE-TFIIH interaction: TFIIE and p53 share the binding region on TFIIH
Okuda, M., Tanaka, A., Satoh, M., Mizuta, S., Takazawa, M., Ohkuma, Y., Nishimura, Y.(2008) EMBO J 27: 1161-1171
- PubMed: 18354501
- DOI: https://doi.org/10.1038/emboj.2008.47
- Primary Citation of Related Structures:
2RNQ, 2RNR - PubMed Abstract:
RNA polymerase II and general transcription factors (GTFs) assemble on a promoter to form a transcription preinitiation complex (PIC). Among the GTFs, TFIIE recruits TFIIH to complete the PIC formation and regulates enzymatic activities of TFIIH. However, the mode of binding between TFIIE and TFIIH is poorly understood. Here, we demonstrate the specific binding of the C-terminal acidic domain (AC-D) of the human TFIIEalpha subunit to the pleckstrin homology domain (PH-D) of the human TFIIH p62 subunit and describe the solution structures of the free and PH-D-bound forms of AC-D. Although the flexible N-terminal acidic tail from AC-D wraps around PH-D, the core domain of AC-D also interacts with PH-D. AC-D employs an entirely novel binding mode, which differs from the amphipathic helix method used by many transcriptional activators. So the binding surface between PH-D and AC-D is much broader than the specific binding surface between PH-D and the p53 acidic fragments. From our in vitro studies, we demonstrate that this interaction could be a switch to replace p53 with TFIIE on TFIIH in transcription.
- Laboratory of Structural Biology, Graduate School of Supramolecular Biology, Yokohama City University, Yokohama, Japan.
Organizational Affiliation: