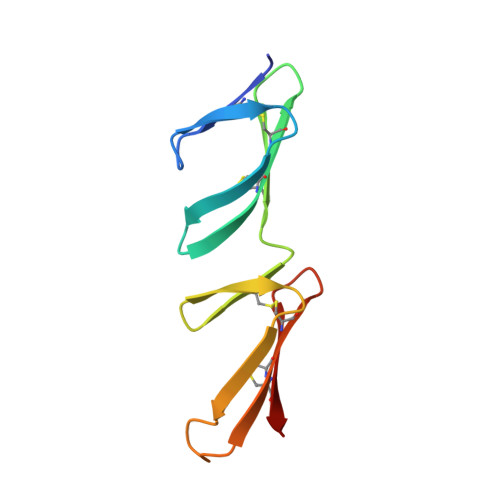
Crystal structures of fibronectin-binding sites from Staphylococcus aureus FnBPA in complex with fibronectin domains
Bingham, R.J., Meenan, N.A., Schwarz-Linek, U., Turkenburg, J.P., Garman, E.F., Potts, J.R.(2008) Proc Natl Acad Sci U S A 105: 12254-12258
- PubMed: 18713862
- DOI: https://doi.org/10.1073/pnas.0803556105
- Primary Citation of Related Structures:
2RKY, 2RKZ, 2RL0, 3CAL - PubMed Abstract:
Staphylococcus aureus can adhere to and invade endothelial cells by binding to the human protein fibronectin (Fn). FnBPA and FnBPB, cell wall-attached proteins from S. aureus, have multiple, intrinsically disordered, high-affinity binding repeats (FnBRs) for Fn. Here, 30 years after the first report of S. aureus/Fn interactions, we present four crystal structures that together comprise the structures of two complete FnBRs, each in complex with four of the N-terminal modules of Fn. Each approximately 40-residue FnBR forms antiparallel strands along the triple-stranded beta-sheets of four sequential F1 modules ((2-5)F1) with each FnBR/(2-5)F1 interface burying a total surface area of approximately 4,300 A(2). The structures reveal the roles of residues conserved between S. aureus and Streptococcus pyogenes FnBRs and show that there are few linker residues between FnBRs. The ability to form large intermolecular interfaces with relatively few residues has been proposed to be a feature of disordered proteins, and S. aureus/Fn interactions provide an unusual illustration of this efficiency.
- Department of Biology, University of York, PO Box 373, York YO10 5YW, United Kingdom.
Organizational Affiliation: