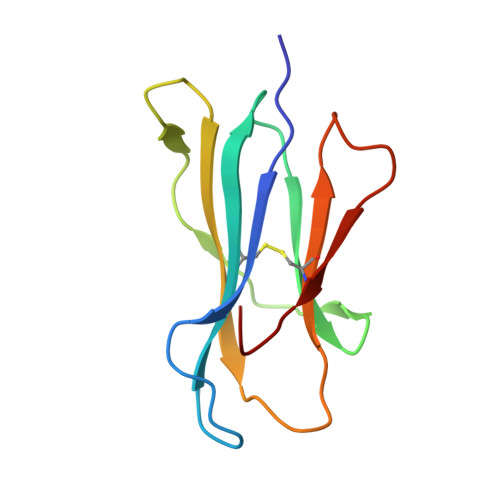
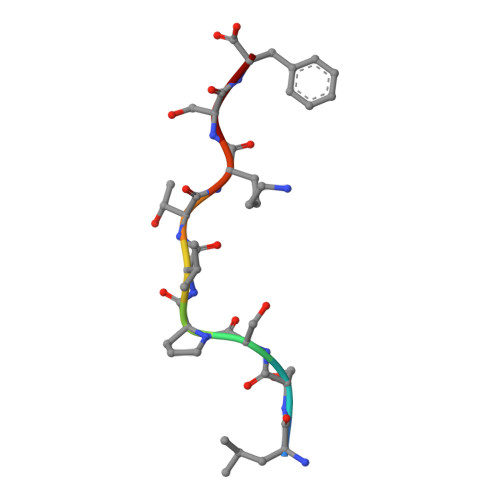
Human leukocyte antigen class I-restricted activation of CD8+ T cells provides the immunogenetic basis of a systemic drug hypersensitivity
Chessman, D., Kostenko, L., Lethborg, T., Purcell, A.W., Williamson, N.A., Chen, Z., Kjer-Nielsen, L., Mifsud, N.A., Tait, B.D., Holdsworth, R., Almeida, C.A., Nolan, D., Macdonald, W.A., Archbold, J.K., Kellerher, A.D., Marriott, D., Mallal, S., Bharadwaj, M., Rossjohn, J., McCluskey, J.(2008) Immunity 28: 822-832
- PubMed: 18549801
- DOI: https://doi.org/10.1016/j.immuni.2008.04.020
- Primary Citation of Related Structures:
2RFX - PubMed Abstract:
The basis for strong immunogenetic associations between particular human leukocyte antigen (HLA) class I allotypes and inflammatory conditions like Behçet's disease (HLA-B51) and ankylosing spondylitis (HLA-B27) remain mysterious. Recently, however, even stronger HLA associations are reported in drug hypersensitivities to the reverse-transcriptase inhibitor abacavir (HLA-B57), the gout prophylactic allopurinol (HLA-B58), and the antiepileptic carbamazepine (HLA-B*1502), providing a defined disease trigger and suggesting a general mechanism for these associations. We show that systemic reactions to abacavir were driven by drug-specific activation of cytokine-producing, cytotoxic CD8+ T cells. Recognition of abacavir required the transporter associated with antigen presentation and tapasin, was fixation sensitive, and was uniquely restricted by HLA-B*5701 and not closely related HLA allotypes with polymorphisms in the antigen-binding cleft. Hence, the strong association of HLA-B*5701 with abacavir hypersensitivity reflects specificity through creation of a unique ligand as well as HLA-restricted antigen presentation, suggesting a basis for the strong HLA class I-association with certain inflammatory disorders.
- Department of Microbiology & Immunology, The University of Melbourne, Parkville, Victoria 3010, Australia.
Organizational Affiliation: