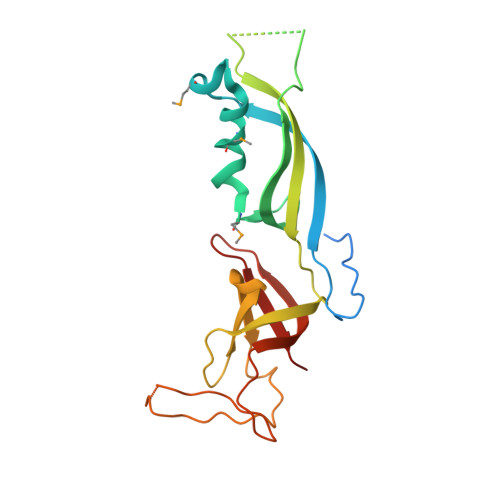
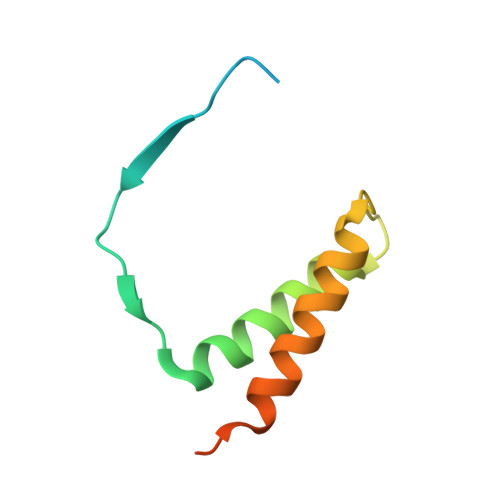
Functional architecture of RNA polymerase I.
Kuhn, C.D., Geiger, S.R., Baumli, S., Gartmann, M., Gerber, J., Jennebach, S., Mielke, T., Tschochner, H., Beckmann, R., Cramer, P.(2007) Cell 131: 1260-1272
- PubMed: 18160037
- DOI: https://doi.org/10.1016/j.cell.2007.10.051
- Primary Citation of Related Structures:
2RF4 - PubMed Abstract:
Synthesis of ribosomal RNA (rRNA) by RNA polymerase (Pol) I is the first step in ribosome biogenesis and a regulatory switch in eukaryotic cell growth. Here we report the 12 A cryo-electron microscopic structure for the complete 14-subunit yeast Pol I, a homology model for the core enzyme, and the crystal structure of the subcomplex A14/43. In the resulting hybrid structure of Pol I, A14/43, the clamp, and the dock domain contribute to a unique surface interacting with promoter-specific initiation factors. The Pol I-specific subunits A49 and A34.5 form a heterodimer near the enzyme funnel that acts as a built-in elongation factor and is related to the Pol II-associated factor TFIIF. In contrast to Pol II, Pol I has a strong intrinsic 3'-RNA cleavage activity, which requires the C-terminal domain of subunit A12.2 and, apparently, enables ribosomal RNA proofreading and 3'-end trimming.
- Gene Center Munich and Center for Integrated Protein Science CIPSM, Department of Chemistry and Biochemistry, Ludwig-Maximilians-Universität München, Feodor-Lynen-Str. 25, 81377 Munich, Germany.
Organizational Affiliation: