Residues substitution in the active site of DSBA may compensate for the lack of the canonical motif CPHC
Rinaldi, F.C., Guimaraes, B.G.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Disulfide oxidoreductase | 193 | Xylella fastidiosa | Mutation(s): 0 Gene Names: XF1436 EC: 1.8.4.2 | 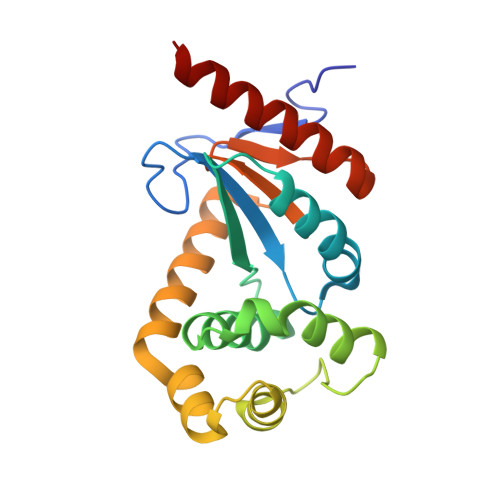 | |
UniProt | |||||
Find proteins for Q9PDE3 (Xylella fastidiosa (strain 9a5c)) Explore Q9PDE3 Go to UniProtKB: Q9PDE3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9PDE3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 8 residue peptide | D [auth T] | 8 | Xylella fastidiosa | Mutation(s): 0 |  |
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 200.117 | α = 90 |
| b = 41.722 | β = 95.87 |
| c = 79.807 | γ = 90 |
| Software Name | Purpose |
|---|---|
| DENZO | data reduction |
| SCALEPACK | data scaling |
| SHARP | phasing |
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |
| CBASS | data collection |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |