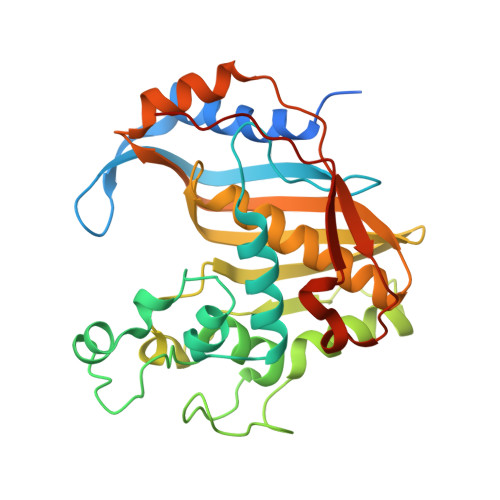
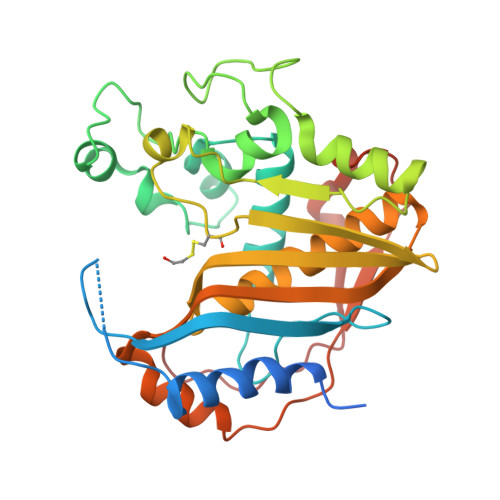
The R163K Mutant of Human Thymidylate Synthase Is Stabilized in an Active Conformation: Structural Asymmetry and Reactivity of Cysteine 195.
Gibson, L.M., Lovelace, L.L., Lebioda, L.(2008) Biochemistry 47: 4636-4643
- PubMed: 18370400
- DOI: https://doi.org/10.1021/bi7019386
- Primary Citation of Related Structures:
2RD8, 2RDA - PubMed Abstract:
Loop 181-197 of human thymidylate synthase (hTS) populates two conformational states. In the first state, Cys195, a residue crucial for catalytic activity, is in the active site (active conformer); in the other conformation, it is about 10 A away, outside the active site (inactive conformer). We have designed and expressed an hTS variant, R163K, in which the inactive conformation is destabilized. The activity of this mutant is 33% higher than that of wt hTS, suggesting that at least one-third of hTS populates the inactive conformer. Crystal structures of R163K in two different crystal forms, with six and two subunits per asymmetric part of the unit cells, have been determined. All subunits of this mutant are in the active conformation while wt hTS crystallizes as the inactive conformer in similar mother liquors. The structures show differences in the environment of catalytic Cys195, which correlate with Cys195 thiol reactivity, as judged by its oxidation state. Calculations show that the molecular electrostatic potential at Cys195 differs between the subunits of the dimer. One of the dimers is asymmetric with a phosphate ion bound in only one of the subunits. In the absence of the phosphate ion, that is in the inhibitor-free enzyme, the tip of loop 47-53 is about 11 A away from the active site.
- Department of Chemistry and Biochemistry and Center for Colon Cancer Research, University of South Carolina, Columbia, SC 29208, USA.
Organizational Affiliation: