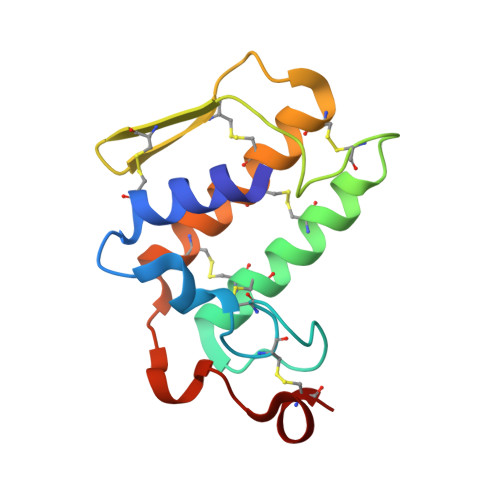
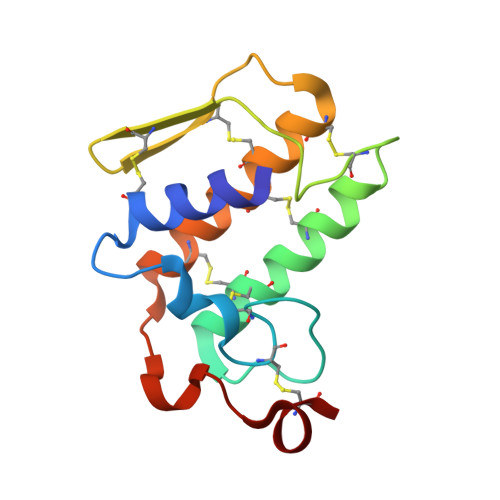
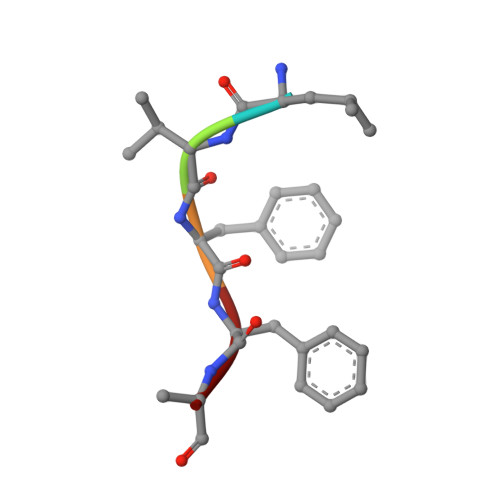
Design of specific inhibitors of Phospholipase A2: Crystal structure of the complex of phospholipase A2 with pentapeptide Leu-Val-Phe-Phe-Ala at 2.9 A resolution
Mirza, Z., Kaur, A., Singh, N., Sinha, M., Sharma, S., Srinivasan, A., Kaur, P., Singh, T.P.To be published.