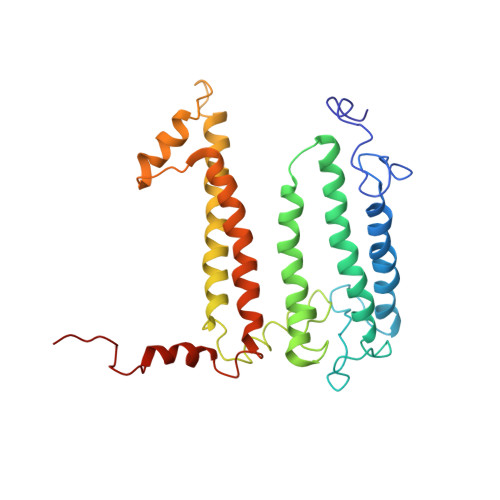
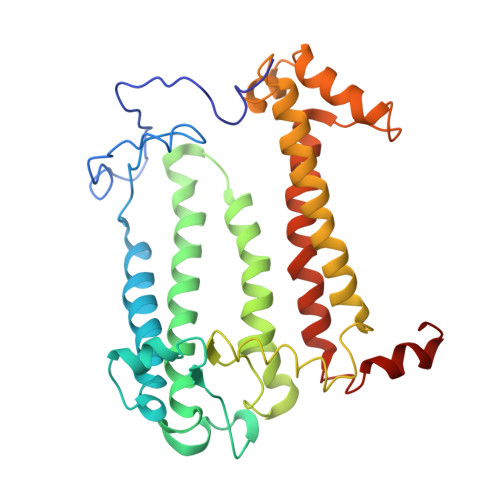
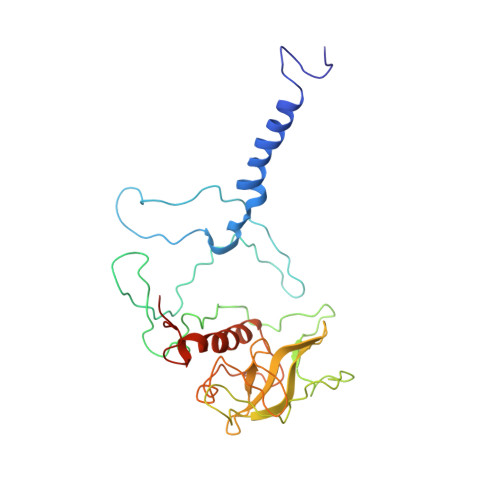
Structure of the membrane-bound protein photosynthetic reaction center from Rhodobacter sphaeroides.
Chang, C.H., el-Kabbani, O., Tiede, D., Norris, J., Schiffer, M.(1991) Biochemistry 30: 5352-5360
- PubMed: 2036404
- DOI: https://doi.org/10.1021/bi00236a005
- Primary Citation of Related Structures:
2RCR - PubMed Abstract:
The structure of the photosynthetic reaction center (RC) from Rhodobacter sphaeroides was determined at 3.1-A resolution by the molecular replacement method, using the Rhodopseudomonas viridis RC as the search structure. Atomic coordinates were refined with the difference Fourier method and restrained least-squares refinement techniques to a current R factor of 22%. The tertiary structure of the RC complex is stabilized by hydrophobic interactions between the L and M chains, by interactions of the pigments with each other and with the L and M chains, by residues from the L and M chains that coordinate to the Fe2+, by salt bridges that are formed between the L and M chains and the H chain, and possibly by electrostatic forces between the ends of helices. The conserved residues at the N-termini of the L and M chains were identified as recognition sites for the H chain.
- Biological and Medical Research Division, Argonne National Laboratory, Illinois 60439.
Organizational Affiliation: