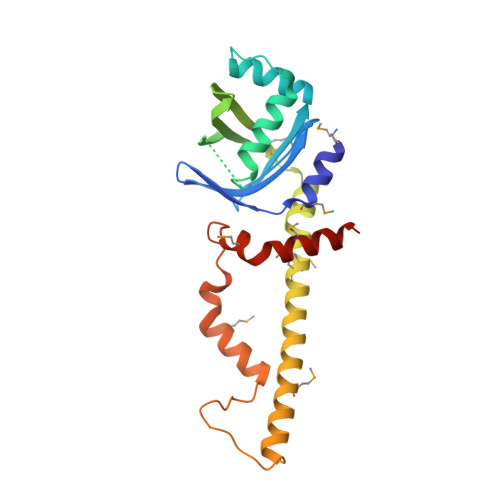
Crystal Structure of Human XLF: A Twist in Nonhomologous DNA End-Joining
Andres, S.N., Modesit, M., Tsai, C.J., Chu, G., Junop, M.S.(null) Mol Cell 28: 1093-1101
- PubMed: 18158905
- DOI: https://doi.org/10.1016/j.molcel.2007.10.024
- Primary Citation of Related Structures:
2R9A - PubMed Abstract:
DNA double-strand breaks represent one of the most severe forms of DNA damage in mammalian cells. One pathway for repairing these breaks occurs via nonhomologous end-joining (NHEJ) and depends on XRCC4, LigaseIV, and Cernunnos, also called XLF. Although XLF stimulates XRCC4/LigaseIV to ligate mismatched and noncohesive DNA ends, the mechanistic basis for this function remains unclear. Here we report the structure of a partially functional 224 residue N-terminal fragment of human XLF. Despite only weak sequence similarity, XLF(1-170) shares structural homology with XRCC4(1-159). However, unlike the highly extended 130 A helical domain observed in XRCC4, XLF adopts a more compact, folded helical C-terminal region involving two turns and a twist, wrapping back to the structurally conserved N terminus. Mutational analysis of XLF and XRCC4 reveals a potential interaction interface, suggesting a mechanism for how XLF stimulates the ligation of mismatched ends.
- Department of Biochemistry and Biomedical Sciences, McMaster University, Hamilton, ON L8N 3Z5, Canada.
Organizational Affiliation: