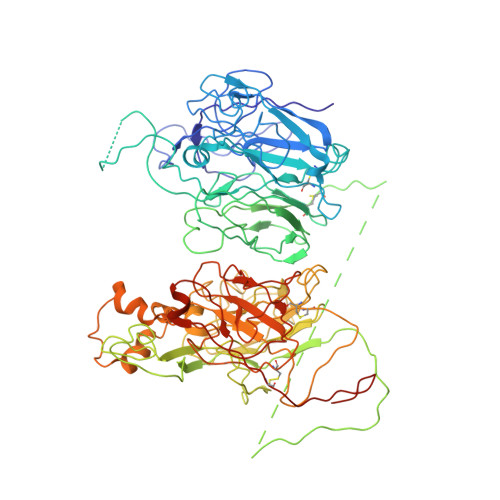
The tertiary structure and domain organization of coagulation factor VIII.
Shen, B.W., Spiegel, P.C., Chang, C.H., Huh, J.W., Lee, J.S., Kim, J., Kim, Y.H., Stoddard, B.L.(2008) Blood 111: 1240-1247
- PubMed: 17965321
- DOI: https://doi.org/10.1182/blood-2007-08-109918
- Primary Citation of Related Structures:
2R7E - PubMed Abstract:
Factor VIII (fVIII) is a serum protein in the coagulation cascade that nucleates the assembly of a membrane-bound protease complex on the surface of activated platelets at the site of a vascular injury. Hemophilia A is caused by a variety of mutations in the factor VIII gene and typically requires replacement therapy with purified protein. We have determined the structure of a fully active, recombinant form of factor VIII (r-fVIII), which consists of a heterodimer of peptides, respectively containing the A1-A2 and A3-C1-C2 domains. The structure permits unambiguous modeling of the relative orientations of the 5 domains of r-fVIII. Comparison of the structures of fVIII, fV, and ceruloplasmin indicates that the location of bound metal ions and of glycosylation, both of which are critical for domain stabilization and association, overlap at some positions but have diverged at others.
- Program in Molecular Biophysics, Structure and Design, Division of Basic Sciences, Fred Hutchinson Cancer Research Center, Seattle, WA, USA.
Organizational Affiliation: