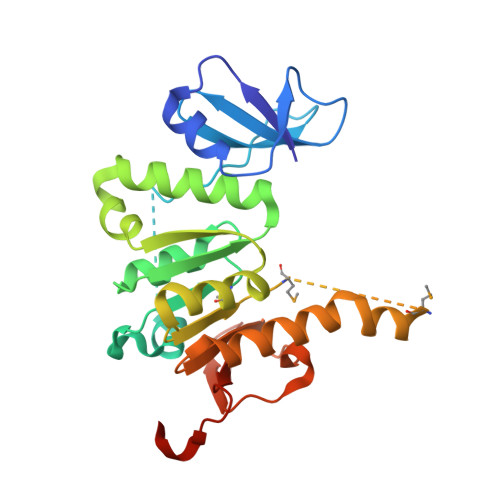
Crystal structure of the SAM-dependent methyltransferase NGO1261 from Neisseria gonorrhoeae.
Forouhar, F., Abashidze, M., Seetharaman, J., Mao, L., Nwosu, C., Fang, Y., Xiao, R., Baran, M.C., Acton, T.B., Montelione, G.T., Tong, L., Hunt, J.F., Northeast Structural Genomics Consortium (NESG)To be published.