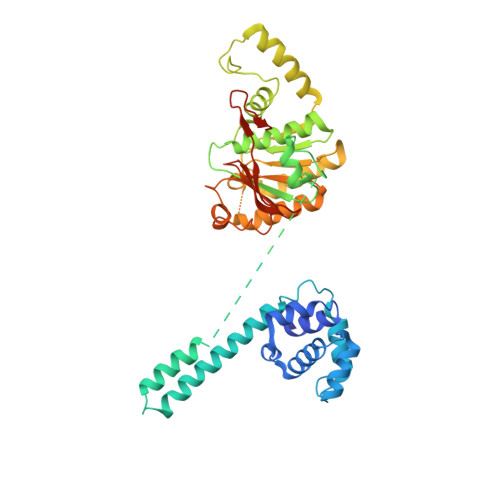
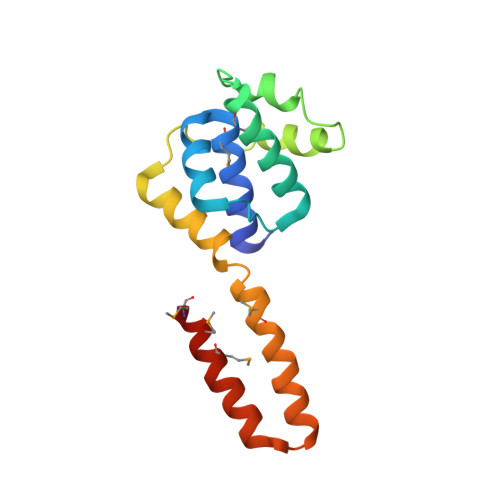
Structure of hexameric DnaB helicase and its complex with a domain of DnaG primase
Bailey, S., Eliason, W.K., Steitz, T.A.(2007) Science 318: 459-463
- PubMed: 17947583
- DOI: https://doi.org/10.1126/science.1147353
- Primary Citation of Related Structures:
2R6A, 2R6C, 2R6D, 2R6E - PubMed Abstract:
The complex between the DnaB helicase and the DnaG primase unwinds duplex DNA at the eubacterial replication fork and synthesizes the Okazaki RNA primers. The crystal structures of hexameric DnaB and its complex with the helicase binding domain (HBD) of DnaG reveal that within the hexamer the two domains of DnaB pack with strikingly different symmetries to form a distinct two-layered ring structure. Each of three bound HBDs stabilizes the DnaB hexamer in a conformation that may increase its processivity. Three positive, conserved electrostatic patches on the N-terminal domain of DnaB may also serve as a binding site for DNA and thereby guide the DNA to a DnaG active site.
- Department of Molecular Biophysics and Biochemistry and Howard Hughes Medical Institute, Yale University, New Haven, CT 06520, USA.
Organizational Affiliation: