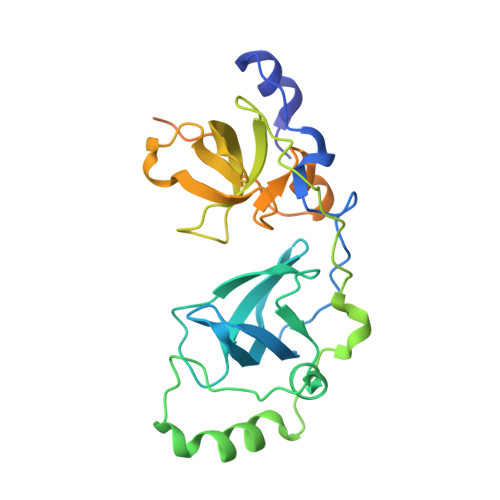
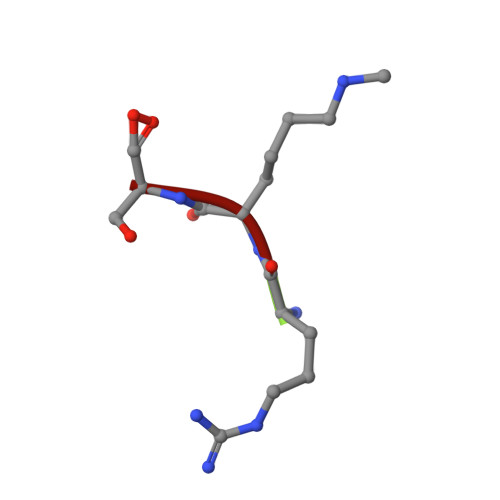
Structural and functional analyses of methyl-lysine binding by the malignant brain tumour repeat protein Sex comb on midleg.
Grimm, C., de Ayala Alonso, A.G., Rybin, V., Steuerwald, U., Ly-Hartig, N., Fischle, W., Muller, J., Muller, C.W.(2007) EMBO Rep 8: 1031-1037
- PubMed: 17932512
- DOI: https://doi.org/10.1038/sj.embor.7401085
- Primary Citation of Related Structures:
2R57, 2R58, 2R5A, 2R5M - PubMed Abstract:
Sex comb on midleg (Scm) is a member of the Polycomb group of proteins involved in the maintenance of repression of Hox and other developmental control genes in Drosophila. The two malignant brain tumour (MBT) repeats of Scm form a domain that preferentially binds to monomethylated lysine residues either as a free amino acid or in the context of peptides, while unmodified or di- or trimethylated lysine residues are bound with significantly lower affinity. The crystal structure of a monomethyl-lysine-containing histone tail peptide bound to the MBT repeat domain shows that the methyl-lysine side chain occupies a binding pocket in the second MBT repeat formed by three conserved aromatic residues and one aspartate. Insertion of the monomethylated side chain into this pocket seems to be the main contributor to the binding affinity. Functional analyses in Drosophila show that the MBT domain of Scm and its methyl-lysine-binding activity are required for repression of Hox genes.
- European Molecular Biology Laboratory, Grenoble Outstation, 6 Rue Jules Horowitz, 38042 Grenoble, France.
Organizational Affiliation: