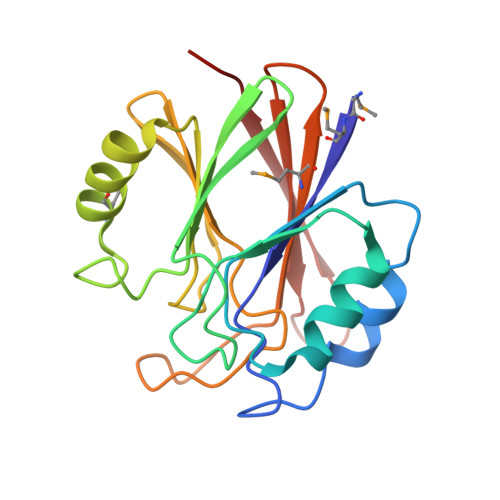
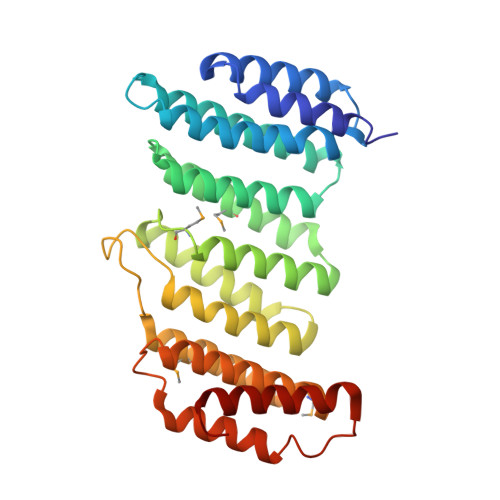
Functional architecture of the retromer cargo-recognition complex.
Hierro, A., Rojas, A.L., Rojas, R., Murthy, N., Effantin, G., Kajava, A.V., Steven, A.C., Bonifacino, J.S., Hurley, J.H.(2007) Nature 449: 1063-1067
- PubMed: 17891154
- DOI: https://doi.org/10.1038/nature06216
- Primary Citation of Related Structures:
2R17 - PubMed Abstract:
The retromer complex is required for the sorting of acid hydrolases to lysosomes, transcytosis of the polymeric immunoglobulin receptor, Wnt gradient formation, iron transporter recycling and processing of the amyloid precursor protein. Human retromer consists of two smaller complexes: the cargo recognition VPS26-VPS29-VPS35 heterotrimer and a membrane-targeting heterodimer or homodimer of SNX1 and/or SNX2 (ref. 13). Here we report the crystal structure of a VPS29-VPS35 subcomplex showing how the metallophosphoesterase-fold subunit VPS29 (refs 14, 15) acts as a scaffold for the carboxy-terminal half of VPS35. VPS35 forms a horseshoe-shaped, right-handed, alpha-helical solenoid, the concave face of which completely covers the metal-binding site of VPS29, whereas the convex face exposes a series of hydrophobic interhelical grooves. Electron microscopy shows that the intact VPS26-VPS29-VPS35 complex is a stick-shaped, flexible structure, approximately 21 nm long. A hybrid structural model derived from crystal structures, electron microscopy, interaction studies and bioinformatics shows that the alpha-solenoid fold extends the full length of VPS35, and that VPS26 is bound at the opposite end from VPS29. This extended structure presents multiple binding sites for the SNX complex and receptor cargo, and appears capable of flexing to conform to curved vesicular membranes.
- Laboratory of Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, US Department of Health and Human Services, Bethesda, Maryland 20892, USA.
Organizational Affiliation: