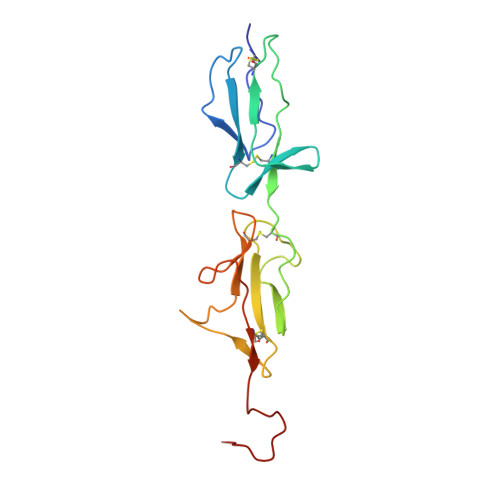
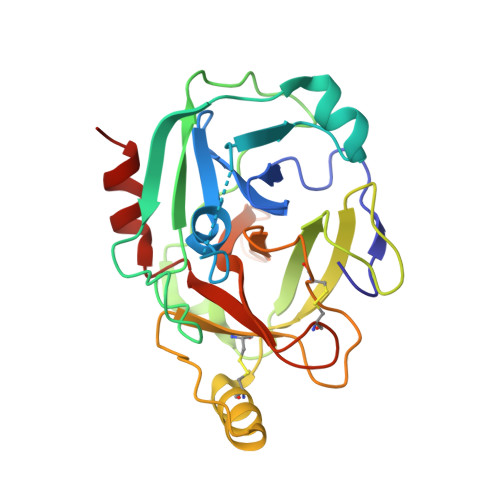
Revisiting the mechanism of the autoactivation of the complement protease C1r in the C1 complex: Structure of the active catalytic region of C1r.
Kardos, J., Harmat, V., Pallo, A., Barabas, O., Szilagyi, K., Graf, L., Naray-Szabo, G., Goto, Y., Zavodszky, P., Gal, P.(2008) Mol Immunol 45: 1752-1760
- PubMed: 17996945
- DOI: https://doi.org/10.1016/j.molimm.2007.09.031
- Primary Citation of Related Structures:
2QY0 - PubMed Abstract:
C1r is a modular serine protease which is the autoactivating component of the C1 complex of the classical pathway of the complement system. We have determined the first crystal structure of the entire active catalytic region of human C1r. This fragment contains the C-terminal serine protease (SP) domain and the preceding two complement control protein (CCP) modules. The activated CCP1-CCP2-SP fragment makes up a dimer in a head-to-tail fashion similarly to the previously characterized zymogen. The present structure shows an increased number of stabilizing interactions. Moreover, in the crystal lattice there is an enzyme-product relationship between the C1r molecules of neighboring dimers. This enzyme-product complex exhibits the crucial S1-P1 salt bridge between Asp631 and Arg446 residues, and intermolecular interaction between the CCP2 module and the SP domain. Based on these novel structural information we propose a new split-and-reassembly model for the autoactivation of the C1r. This model is consistent with experimental results that have not been explained adequately by previous models. It allows autoactivation of C1r without large-scale, directed movement of C1q arms. The model is concordant with the stability of the C1 complex during activation of the next complement components.
- Department of Biochemistry, Institute of Biology, Eötvös Loránd University, Pázmány sétány 1/C, Budapest H-1117, Hungary.
Organizational Affiliation: