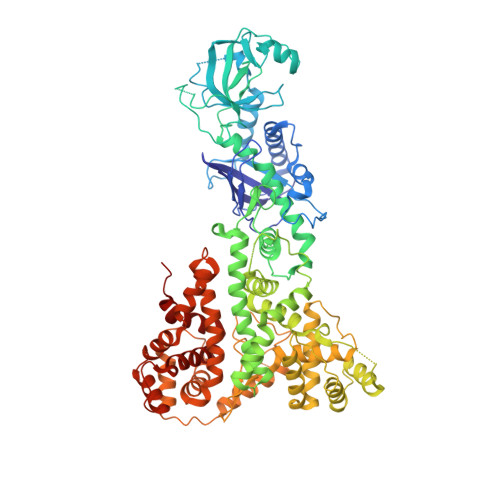
An unusual case of pseudo-merohedral twinning in orthorhombic crystals of Dicer
Macrae, I.J., Doudna, J.A.(2007) Acta Crystallogr D Biol Crystallogr 63: 993-999
- PubMed: 17704568
- DOI: https://doi.org/10.1107/S0907444907036128
- Primary Citation of Related Structures:
2QVW - PubMed Abstract:
The crystal structure of the enzyme Dicer from Giardia intestinalis was solved to 3.3 A resolution by MAD using crystals belonging to space group P2(1)2(1)2 [Macrae et al. (2006), Science, 311, 195-198]. These crystals were derived from crystals that diffracted X-rays to 3.0 A resolution but were refractory to structure determination because they were twinned. It is shown here that the original Dicer crystals represent an unusual case of perfect pseudo-merohedral twinning of orthorhombic crystals. Before the twinning problem was overcome, it was possible to calculate a low-resolution electron-density map in space group P4(1) that was used to build a partial molecular model. Experimental phases were sufficient to identify heavy-atom sites that indicated space-group inconsistency, leading to identification of the true space group. This information guided the search for different crystallization conditions that yielded untwinned crystals and ultimately a fully interpretable electron-density map.
- Howard Hughes Medical Institute, Department of Molecular and Cell Biology, University of California, Berkeley, CA 94720, USA.
Organizational Affiliation: