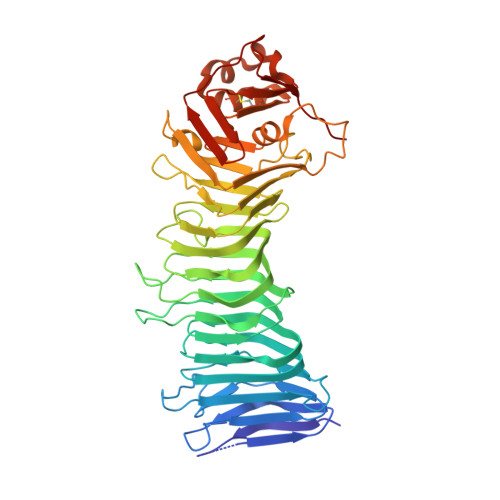
Crystal structure of the Helicobacter pylori vacuolating toxin p55 domain
Gangwer, K.A., Mushrush, D.J., Stauff, D.L., Spiller, B., McClain, M.S., Cover, T.L., Lacy, D.B.(2007) Proc Natl Acad Sci U S A 104: 16293-16298
- PubMed: 17911250
- DOI: https://doi.org/10.1073/pnas.0707447104
- Primary Citation of Related Structures:
2QV3 - PubMed Abstract:
Helicobacter pylori VacA, a pore-forming toxin secreted by an autotransporter pathway, causes multiple alterations in human cells, contributes to the pathogenesis of peptic ulcer disease and gastric cancer, and is a candidate antigen for inclusion in an H. pylori vaccine. Here, we present a 2.4-A crystal structure of the VacA p55 domain, which has an important role in mediating VacA binding to host cells. The structure is predominantly a right-handed parallel beta-helix, a feature that is characteristic of autotransporter passenger domains but unique among known bacterial protein toxins. Notable features of VacA p55 include disruptions in beta-sheet contacts that result in five beta-helix subdomains and a C-terminal domain that contains a disulfide bond. Analysis of VacA protein sequences from unrelated H. pylori strains, including m1 and m2 forms of VacA, allows us to identify structural features of the VacA surface that may be important for interactions with host receptors. Docking of the p55 structure into a 19-A cryo-EM map of a VacA dodecamer allows us to propose a model for how VacA monomers assemble into oligomeric structures capable of membrane channel formation.
- Department of Microbiology and Immunology, Vanderbilt University Medical Center, Nashville, TN 37232, USA.
Organizational Affiliation: